after following Stefan's very helpful answer to this post, where he uses ggnewscale::new_scale(), I now am stuck with the following question:
"How to arrange the custom legends from ggnewscale into multiple vertical columns?"
like it is usually done with a command such as guides(scale_shape_manual=guide_legend(ncol=2)) in ggplot2.
Minimal reproducible example:
# fictional data in the scheme of https://stackoverflow.com/q/66804487/16642045
mutated <- list()
for(i in 1:10) {
mutated[[i]] <- data.frame(Biological.Replicate = rep(1,4),
Reagent.Conc = c(10000, 2500, 625, 156.3),
Reagent = rep(1,8),
Cell.type = rep(LETTERS[i],4),
Mean.Viable.Cells.1 = rep(runif(n = 10, min = 0, max = 1),4))
}
mutated <- do.call(rbind.data.frame, mutated)
The modified code after the answer of user "stefan" looks like this:
# from https://stackoverflow.com/a/66808609/16642045
library(ggplot2)
library(ggnewscale)
library(dplyr)
library(magrittr)
mutated <- mutated %>%
mutate(Cell.type = as.factor(Cell.type),
Reagent = factor(Reagent,
levels = c("0", "1", "2")))
mean_mutated <- mutated %>%
group_by(Reagent, Reagent.Conc, Cell.type) %>%
split(.$Cell.type)
layer_geom_scale <- function(Cell.type) {
list(geom_point(mean_mutated[[Cell.type]], mapping = aes(shape = Reagent)),
geom_line(mean_mutated[[Cell.type]], mapping = aes(group = Reagent, linetype = Reagent)),
scale_linetype_manual(name = Cell.type, values = c("solid", "dashed", "dotted"), drop=FALSE),
scale_shape_manual(name = Cell.type, values = c(15, 16, 4), labels = c("0", "1", "2"), drop=FALSE)
)
}
my_plot <-
ggplot(mapping = aes(
x = as.factor(Reagent.Conc),
y = Mean.Viable.Cells.1)) +
layer_geom_scale(unique(mutated$Cell.type)[1])
for(current_Cell.type_index in 2:length(unique(mutated$Cell.type))) {
my_plot <-
my_plot +
ggnewscale::new_scale("shape") +
ggnewscale::new_scale("linetype") +
layer_geom_scale(unique(mutated$Cell.type)[current_Cell.type_index])
}
my_plot
Now, I want the legends to be displayed side-by-side, in two columns, and I tried this (without success):
my_plot +
guides(scale_shape_manual=guide_legend(ncol=2))
EDIT: A picture of the way I want the legends to be arranged
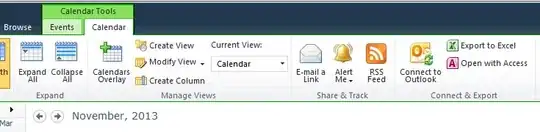
Is there anyone who could help me?
Thanks!