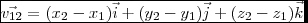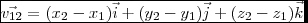EDIT (original, incorrect answer below)
Here's another try at this. As far as I can tell this is a consequence of how the Venn diagram is being constructed. d@region is an object consisting of 15 polygons; ggplot is then looking for a color for each of them when we call geom_sf(aes(fill = name), data = venn_region(d)).
Here's an imperfect try at a solution. This duplicates the venn object (d2 <- process_data(venn)) and then creates polygons from the outlines of the shapes (d2@region <- st_polygonize(d@setEdge)). Those polygons are then used to plot the circles/fill.
library(ggVennDiagram)
library(ggplot2)
library(sf)
genes <- paste0("gene",1:40)
gene_list <- list(
A = genes[1:10],
B = genes[1:20],
C = genes[5:15],
D = genes[30:40]
)
venn <- Venn(gene_list)
d <- process_data(venn)
d2 <- process_data(venn)
d2@region <- st_polygonize(d@setEdge)
col <- c(A = 'blue', B = 'red', C = 'green', D = 'orange')
ggplot() +
geom_sf(aes(fill = name), data = venn_region(d2)) +
geom_sf(aes(color = name), data = venn_setedge(d)) +
geom_sf_text(aes(label = name), data = venn_setlabel(d)) +
geom_sf_text(aes(label = count), data = venn_region(d)) +
scale_color_manual(values = alpha(col, .2)) +
scale_fill_manual(values = alpha(col, .2)) +
theme_void()
I'm not getting a gray color for the intersection region, but instead a message that points to scale_fill_manual requiring more than 4 color values.
Following the answer to this StackOverflow post on creating a color gradient, it's possible to set a color gradient for the colors you provided. Those then get used to fill the regions of intersection, and the 4 colors are the outlines of the regions.
library(ggVennDiagram)
library(ggplot2)
genes <- paste0("gene",1:40)
gene_list <- list(
A = genes[1:10],
B = genes[1:20],
C = genes[5:15],
D = genes[30:40]
)
venn <- Venn(gene_list)
d <- process_data(venn)
# vector for colors
colorGroups <- c(A = 'blue', B = 'red', C = 'green', D = 'orange')
# use colorRampPalette to create function that interpolates colors
colfunc <- colorRampPalette(colorGroups)
# call function and create vector of 15 colors
col <- colfunc(15)
ggplot() +
geom_sf(aes(fill = name), data = venn_region(d)) +
geom_sf(aes(color = name), data = venn_setedge(d)) +
geom_sf_text(aes(label = name), data = venn_setlabel(d)) +
geom_sf_text(aes(label = count), data = venn_region(d)) +
scale_fill_manual(values = alpha(col, .2)) +
scale_color_manual(values = colorGroups) +
theme_void()