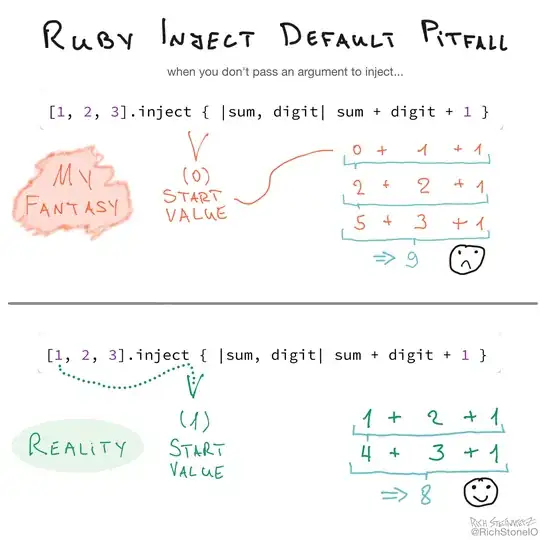
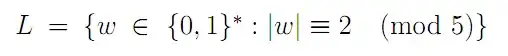I have been generating filled KDEs using sns.kdeplot. To try to better understand what's going on under the hood, I decided to try estimating the KDE myself and plotting the kde using plt.contourf, using code I found in the answers to this question. However, my results using the two methods look quite different. I've tried to make sure my implementation of the KDE estimation matches the way it's done in Seaborn, and I can't find any discrepancies. Can someone help me understand why my plots do not look the same?
Here is some example code:
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
geyser = sns.load_dataset("geyser")
dur = np.array(geyser["duration"])
wait = np.array(geyser["waiting"])
# plot the KDE using sns.kdeplot
sns.kdeplot(dur, wait, shade=True, cmap="binary", cut=0)
# calculate my own KDE and plot using plt.contourf
def make_kde(x, y):
"""
Based on
https://stackoverflow.com/questions/50917216/log-scales-with-seaborn-kdeplot
"""
kde = stats.gaussian_kde([x, y])
xx, yy = np.mgrid[min(x):max(x):(max(x)-min(x))/200,
min(y):max(y):(max(y)-min(y))/200]
density = kde(np.c_[xx.flat, yy.flat].T).reshape(xx.shape)
return xx, yy, density
xgrid, ygrid, density = make_kde(dur, wait)
fig, ax = plt.subplots()
ax.contourf(xgrid, ygrid, density, cmap="binary", levels=10)
Here is the plot generated by sns.kdeplot:
And here is the plot generated by plt.contourf:
I would really appreciate any tips for trying to understand why these two plots look different.