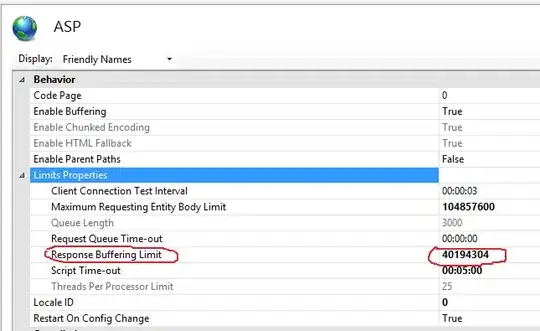
Your main issue is that you misunderstand what .read_adjlist does. It expects the graph to be described using adjacency lists. It sees a row starting with "Step 1", so it thinks it means "Step 1 is a node that is adjacent to all the other nodes on that row".
At this point, you have two options:
- Modify you datafile so that it is a correct adjacency list representation of your graph;
- Modify your code to read the datafile and interpret it the way you want.
Note that .read_adjlist is not the only way to create a graph. You can use for instance .add_edges_from.
A solution without modifying the datafile would be:
import networkx as nx
import csv
import matplotlib.pyplot as plt
g = nx.Graph()
with open('datatest.csv') as f:
r = csv.reader(f)
g.add_edges_from([(x,y) for row in r for x,y in zip(row, row[1:])])
nx.draw(g)
plt.show() # note it's plt.show(), not just plt.show