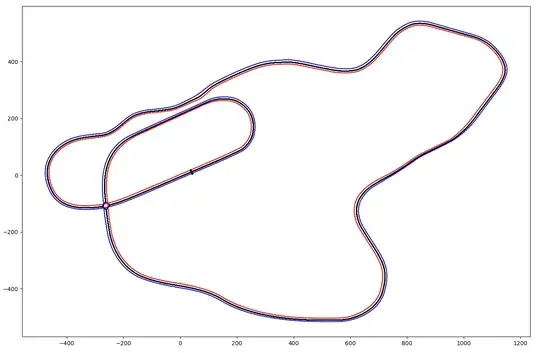
Suppose I have two closed loops representing either boundary of a racetrack in two dimensional x,y space. The start of each loop if connected by a line would represent the start/finish line, the midpoint of this line would represent the centerline of the track.
Moving outward from the start line each loop is comprised of line segments which have random lengths with relatively small variations. The number of line segments is high at approximately 20-40k for each loop.
With this in mind the problem I am trying to solve is finding a closed loop representing the midline of the two other loops (ie - the centerline of the track).
At first I thought given that each small line segment is relatively close in length that I could take a step one segment in length which is the smaller of the two segments of either loop, this shorter length is used to inform a point along the longer segment and then a midpoint is found between the two steps of equal length. The new position along each line is updated and the process repeats effectively ping-ponging back and forth between the active loop having the shorter step forward.
I am doing this in python where I have a csv file containing the x and y position (x = *.PositionX and y = *.PositionZ) for the two loops. For reference below is what I was trying to use
import traceback
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
fDict = {"Bernese Alps":{"fName":"2021-09-12202910_FM7_DASH","left":4,"right":6},
"Maple Valley":{"fName":"2021-08-30171416_FM7_DASH","left":4,"right":1},}
track = "Maple Valley"
df = pd.read_csv(fDict[track]["fName"]+".csv")
#The laps which correspond to track limits right and left
lapRight = fDict[track]["right"]
lapLeft = fDict[track]["left"]
#Position from history for right and left
rightPos = df.loc[df.LapNumber==lapRight,['PositionX','PositionZ']]
leftPos = df.loc[df.LapNumber==lapLeft,['PositionX','PositionZ']]
#Vector difference from one position to the next both right and left
rightDiff = rightPos.diff()
rightDiff.iloc[0] = rightPos.iloc[0]-rightPos.iloc[-1]
leftDiff = leftPos.diff()
leftDiff.iloc[0] = leftPos.iloc[0]-leftPos.iloc[-1]
#Vector difference magnitudes one position to the next both right and left
rightDiffMag = (rightDiff.PositionX.copy()*0+np.linalg.norm(rightDiff,axis=1)).rename({'PositionX':'PositionMag'})
leftDiffMag = (leftDiff.PositionX.copy()*0+np.linalg.norm(leftDiff,axis=1)).rename({'PositionX':'PositionMag'})
#Normalized vectors to project onto
rightDiffNormed = rightDiff.div(rightDiffMag,'index')
leftDiffNormed = leftDiff.div(leftDiffMag,'index')
#Methods to find center point
#(originally avg of x and y was used but change to confirm I was not doing that wrong)
def right_to_left(rightX,rightZ,leftX,leftZ):
rtl_diff = np.array([leftX-rightX,leftZ-rightZ])
rtl_mag = np.sqrt(sum(rtl_diff**2))
rtl_dir = rtl_diff/rtl_mag
return rtl_dir,rtl_mag
def midPoint(rightX,rightZ,leftX,leftZ):
rtl_dir,rtl_mag = right_to_left(rightX,rightZ,leftX,leftZ)
mid_vec = rtl_dir*rtl_mag*.5
return rightX+mid_vec[0],rightZ+mid_vec[1]
#Create new right,left, and mid line containers
rights = [[rightPos.iloc[0].values[0]],[rightPos.iloc[0].values[1]]]
lefts = [[leftPos.iloc[0].values[0]],[leftPos.iloc[0].values[1]]]
midP = midPoint(rights[0][0],rights[1][0],lefts[0][0],lefts[1][0])
mids = [[midP[0]],[midP[1]]]
#Initialize which size is longer
#Initialize partial remainders from prior step
partials = {'left':0.0,'right':0.0}
#Initialize indicie tracking for each side
indicies = {'left':1,'right':1}
print(len(leftPos),len(rightPos))
key = input("PAUSED - 'Y' TO CONTINUE 'N' TO ABORT: ")
if key.lower() == 'y':
Done = False
try:
while not Done:
rightIdx = rightPos.index[indicies['right']]
leftIdx = leftPos.index[indicies['left']]
#Project forward one point
rightIdx_fwd = rightPos.index[0] if indicies['right']>=len(rightPos)-1 else rightPos.index[indicies['right']+1]
leftIdx_fwd = rightPos.index[0] if indicies['left']>=len(leftPos)-1 else leftPos.index[indicies['left']+1]
#If remaining left side is longer
if leftDiffMag.loc[leftIdx]-partials['left'] >= rightDiffMag.loc[rightIdx]-partials['right']:
active_side = 'left'
#Step magnitude is full right step
mag = rightDiffMag.loc[rightIdx]-partials['right']
if not mag+partials['left'] >= leftDiffMag.loc[leftIdx_fwd]:
partials['left']+= mag
else:
partials['left']=0.0
indicies['left']+=1
partials['right']=0.0
indicies['right']+=1
else:
#Step magnitude is full right step
active_side = 'right'
mag = leftDiffMag.loc[leftIdx]-partials['left']
if not mag+partials['right'] >= rightDiffMag.loc[rightIdx_fwd]:
partials['right']+= mag
else:
partials['right']=0.0
indicies['right']+=1
partials['left']=0.0
indicies['left']+=1
rightUpdate = rightPos.loc[rightIdx]+mag*rightDiffNormed.loc[rightIdx_fwd]
leftUpdate = leftPos.loc[leftIdx]+mag*leftDiffNormed.loc[leftIdx_fwd]
midP = midPoint(*rightUpdate.values,*leftUpdate.values)
rights[0].append(rightUpdate.PositionX)
rights[1].append(rightUpdate.PositionZ)
lefts[0].append(leftUpdate.PositionX)
lefts[1].append(leftUpdate.PositionZ)
mids[0].append(midP[0])
mids[1].append(midP[1])
#print(active_side,indicies['left'],leftIdx,indicies['right'],rightIdx,mag)
if indicies['right']>=len(rightPos)-1 and indicies['left']>=len(leftPos)-1:
Done=True
except Exception as err:
traceback.print_exception(None, err, err.__traceback__)
print(len(rights[0]),len(lefts[0]),len(mids[0]))
axi = plt.subplot()
axi.plot(*rights,ls='-',marker='.',c='r')
axi.plot(*lefts,ls='-',marker='.',c='b')
axi.plot(*mids,ls='-',marker='.',c='k')
axi.set_aspect('equal')
fig,axes = plt.subplots(2)
for i in range(2):
axes[i].plot(rights[i],c='r')
axes[i].plot(lefts[i],c='b')
axes[i].plot(mids[i],c='k')
#fig,axi = plt.subplots()
#for rX,rZ,lX,lZ,mX,mZ in zip(*rights,*lefts,*mids):
#axi.plot([rX,mX,lX],[rZ,mZ,lZ],c='k')
finally:
plt.show()
else:
print("Processing was ABORTED!")
This worked better than I expected but:
- It is slow, and
- Depending on rate of curvature/difference in curvature (I am assuming that is what is driving the error) between the two loops over a given turn results in pulling the midline to one side or the other.
- Also, I am not sure if it is an implementation issue or simplify an artifact of the crude method but the update to the centerline is noisy/jagged (see below comment I believe this is an error in the implementation)
I am looking to find any method which will find the centerline. Bonus points if it lends itself well to obtaining a vector normal to the centerline (and its opposite -- rotated 180 degrees) and corresponding intersection of this normal with either of the bounding loops (ie - a line representing midline distance from start at any point along the track). Further bonus points if centerline step size is normalized.
I was thinking one solution, given that I know the direction of travel and center point at the start of both loops, would be to define a step size for the centerline, then use this to represent a half circle with radius equal the step size, then solve for the shortest line tangent to this circle which is bounded by intersection with the two outer loops. I have crudely illustrated this below. I am not sure there is an efficient/elegant way to solve using this method.
Thank you for any feedback/direction you can provide. I think there is probably a known solution to this type of problem and I am simply using the wrong search terms trying to find examples.
PS - Looking more closely and it appears there are aberrations in the update to the outer loops which likely helps explain the jagged update of the midline. Regardless I think the skew of the centerline to one side or the other remains a shortcoming of this approach and not an implementation problem.
Update: It would appear this is frequently a problem in the GIS domain where polylines are used to represent the boundaries of for instance a river and the rivers midline is desired.
Apparently one solution is to generate a Voronoi diagram. Doing this will naturally produce poly vertices and edges representing the midline.
Example: https://gis.stackexchange.com/questions/269238/create-a-center-line-between-2-polyline-qgis
I did a quick test of this following the documentation for scipy.spatial.voronoi_plot_2d (https://docs.scipy.org/doc/scipy/reference/generated/scipy.spatial.voronoi_plot_2d.html) and it worked well even with the 47k+ points.
Now I just need to detect those Voronoi polygon verticies which are inside the two outer boundaries.
Additional word of caution; As you might expect this method has a hard time with the self intersecting track and so I will probably have to detect this condition or manually prevent it by cutting the track into multiple parts then sewing them back together.