The underlying axes-level functions are sns.regplot and sns.residplot. The example data used in the question's post seems to use y-values which are close to normal distributed around zero, which makes it rather hard to see what is happening.
The code below uses the Gentoo species of the penguins dataset.
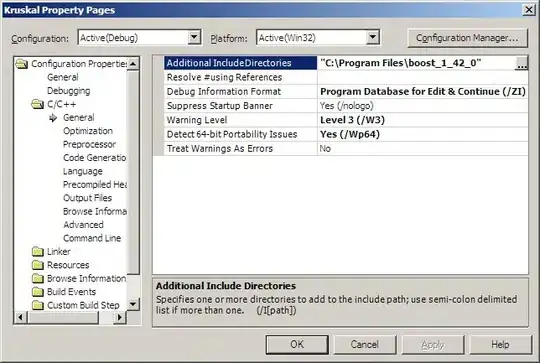
The regplot at the left shows the original data with a regression line. The y-values are all positive, and the regression line has an upward slope.
The residplot at the right shows the data with the regression line subtracted. The y-values are positive where the original data are above the regression line, and negative below. The residplot helps to assess whether the regression (linear in this case) is a suitable fit.
The linked tutorial post and video might be helpful to better understand the residual plot.
import matplotlib.pyplot as plt
import seaborn as sns
penguins = sns.load_dataset('penguins')
penguins = penguins[penguins['species'] == 'Gentoo']
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(14, 4), sharex=True)
sns.regplot(data=penguins, x="bill_length_mm", y="bill_depth_mm", ax=ax1)
ax1.set_title("regplot")
sns.residplot(data=penguins, x="bill_length_mm", y="bill_depth_mm", ax=ax2)
ax2.set_title("residplot")
plt.tight_layout()
plt.show()
If your input data are already residuals, both plots seem to be very similar:
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
penguins = sns.load_dataset('penguins').dropna()
penguins = penguins[penguins['species'] == 'Gentoo']
x = penguins['bill_length_mm']
y = penguins['bill_depth_mm']
regr = stats.linregress(x, y)
y -= regr.slope * x + regr.intercept # calculate the residuals
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(15, 4))
sns.regplot(x=x, y=y, lowess=True, ax=ax1)
ax1.set_title('regplot')
sns.residplot(x=x, y=y, lowess=True, ax=ax2)
ax2.set_title('residplot')
plt.tight_layout()
plt.show()