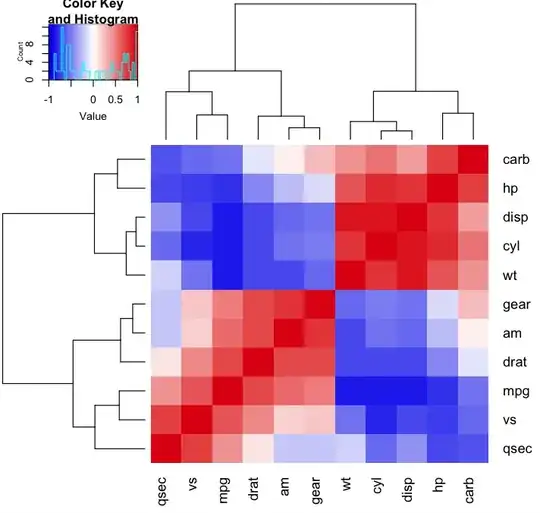
Try using geom_segment.
graph <- graph[complete.cases(graph),]
graph$time_end <- lead(graph$Time, n=1)
graph$HR_end <- lead(graph$HR, n = 1)
####Creating figure####
graph$intensity<-factor(graph$Intensity,
levels =c("Sedentary", "Light", "Moderate", "Vigorous"))
ggplot(graph, aes(x = Time, y = HR)) +
geom_segment(aes(xend = time_end, yend = HR_end, color = intensity)) +
labs(x = "Time", y = "Heart Rate (bpm)")+
scale_color_discrete("Intensity:", labels = c("Sedentary", "Light",
"Moderate", "Vigorous")) +
theme(legend.title = element_text(family = "Playfair",
color = "chocolate",
size = 10, face = 2))+
theme(panel.grid = element_blank()) +
geom_rug(show.legend = FALSE)+
scale_x_datetime(date_breaks = "4 hours", date_labels = "%H:%M")+
ylim(c(0,160))+
scale_y_continuous(breaks = seq(0, 160, by =40))
Is this what you are looking for?
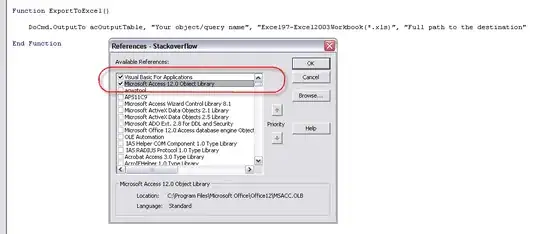
As tjebo noted, there is still some overlap in the lines. I understand, though, that there is a specific aesthetic you are looking to achieve, and I'd like a cigar. If we go back to your original solution, geom_line() didn't work because it creates a continuous line for each factor, in this case the Intensity. I suggested you use geom_segment(). With this, you define the start and end points and can determine the color of the line. The problem here is that the points fall into one zone or another, but the lines may cross over the zone/intensities. That is why my early graph had overlaps. If you think it is appropriate, you need to add more points to your data to account for when the thresholds are crossed. Essentially, it adds two new points that are just at and over the threshold. So I've done that in the code below and have gotten you closer. The problem I didn't solve for are the cases when a zone is skipped, for example, it goes from zone 1/sedentary to zone 3/moderate. So you'll note there appear some small overlaps. That could be worked out, but will leave that to you.
library(tidyverse)
library(hms)
library(dplyr)
library(lubridate)
library(ggplot2)
#Import dataset#
graph <-read.csv("graph.csv", sep=";", na.strings = "?")
graph$HR <- as.numeric(as.character(graph$HR))
graph$Zone <- as.numeric(as.character(graph$Zone))
graph$Time =as.POSIXct(graph$Time,format="%H:%M:%S")
#graph$Time = as_hms(graph$Time) This throws off scale_x_datetime() so didn't use
#remove NA values
graph <- graph[complete.cases(graph),]
#understand zones
graph %>%
group_by(Zone) %>%
summarise(min = min(HR), max = max(HR))
#create Heart Rate Theshold values
thresholds <- tibble("Zone" = c(1, 2, 3, 4), "HRT" = c(78, 97, 116, Inf))
graph <- merge(thresholds, graph, by = "Zone")
graph <- graph[order(graph$Time),]
#generate new data points for instances the Zone is crossed
#note we introduce a new time point that is between the two original times
new_points <- tibble()
for(r in 1:(nrow(graph)-1)){
if(graph[r,"Zone"] == graph[r+1, "Zone"]){
next
} else if(graph[r,"Zone"] < graph[r+1, "Zone"]) {
new <- tibble(Zone = c(graph[r,"Zone"], graph[r+1, "Zone"]),
Time = c(graph[r,"Time"] + 0.5, graph[r+1, "Time"] - 0.4),
HR = c(graph[r, "HRT"], graph[r, "HRT"] + 1),
Intensity = c(graph[r,"Intensity"], graph[r+1, "Intensity"]))
} else if(graph[r,"Zone"] > graph[r+1, "Zone"]) {
new <- tibble(Zone = c(graph[r,"Zone"], graph[r+1, "Zone"]),
Time = c(graph[r,"Time"] + 0.5, graph[r+1, "Time"] - 0.4),
HR = c(graph[r+1, "HRT"] +1 , graph[r+1, "HRT"]),
Intensity = c(graph[r,"Intensity"], graph[r+1, "Intensity"]))
}
new_points <- rbind(new_points, new)
}
new_points <- merge(thresholds, new_points, by = "Zone")
#merge and reorder
graph <- rbind(graph, new_points)
graph <- graph[order(graph$Time),]
#create end points for segments
graph$time_end <- lead(graph$Time, n=1)
graph$HR_end <- lead(graph$HR, n = 1)
####Creating figure####
graph$intensity<-factor(graph$Intensity,
levels =c("Sedentary", "Light", "Moderate", "Vigorous"))
ggplot(graph, aes(x = Time, y = HR)) +
geom_segment(aes(xend = time_end, yend = HR_end, color = intensity)) +
labs(x = "Time", y = "Heart Rate (bpm)")+
scale_color_discrete("Intensity:", labels = c("Sedentary", "Light",
"Moderate", "Vigorous")) +
theme(legend.title = element_text(#family = "Playfair",
color = "chocolate",
size = 10, face = 2))+
theme(panel.grid = element_blank()) +
geom_rug(show.legend = FALSE, aes(color = intensity))+
scale_x_datetime(date_breaks = "4 hours", date_labels = "%H:%M")+
ylim(c(0,160))+
scale_y_continuous(breaks = seq(0, 160, by =40))