I have timeseries (datetime, Instance, Value) with some NAs in Values. If Value for all Instance - NA for same datetime, that means gap in data collection. I need to highlight that periods.
My example script and data:
library(tidyr)
library(ggplot2)
example.data1 <- data.frame( Instance = rep("A",11),
datetime = seq.POSIXt(as.POSIXct("2020-12-26 10:00:00"), as.POSIXct("2020-12-26 10:00:00") + 15*10, "15 sec"),
Value = c(0,1,2,3,4,5,6,NA,NA,9,10)
)
example.data2 <- data.frame( Instance = rep("B",11),
datetime = seq.POSIXt(as.POSIXct("2020-12-26 10:00:00"), as.POSIXct("2020-12-26 10:00:00") + 15*10, "15 sec"),
Value = c(1,2,NA,4,5,6,7,NA,NA,10,11)
)
example.data3 <- data.frame( Instance = rep("C",11),
datetime = seq.POSIXt(as.POSIXct("2020-12-26 10:00:00"), as.POSIXct("2020-12-26 10:00:00") + 15*10, "15 sec"),
Value = c(2,3,4,5,NA,7,8,NA,NA,11,12)
)
example.data <- bind_rows(example.data1, example.data2, example.data3)
ggplot (data = example.data, aes(x=datetime,y=Value, color = Instance)) +
geom_line(size = 1.2) +
theme_bw()
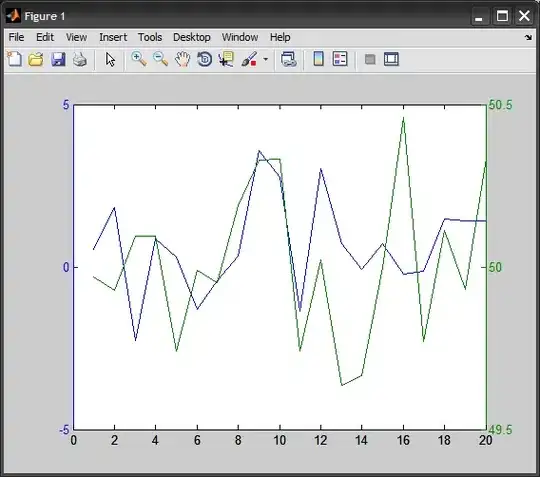
My result picture:
What I really need:
How to reach that?
UPD.
Code is answer below doesn't work correctly. Look at that:
example.data.gap <- example.data %>%
group_by(datetime) %>%
summarise(is_gap = all(is.na(Value))) %>%
# Start and End
mutate(xmin = lag(datetime), xmax = lead(datetime)) %>%
filter(is_gap)
Result is 2 overlapping intervals instead of 1:
# A tibble: 2 x 4
datetime is_gap xmin xmax
<dttm> <lgl> <dttm> <dttm>
1 2020-12-26 10:01:45 TRUE 2020-12-26 10:01:30 2020-12-26 10:02:00
2 2020-12-26 10:02:00 TRUE 2020-12-26 10:01:45 2020-12-26 10:02:15
Picture - we can see that overlaps if we use alpha:
ggplot(data = example.data, aes(x = datetime, y = Value, color = Instance)) +
geom_line(size = 1.2) +
geom_rect(data = example.data.gap, aes(xmin = xmin, xmax = xmax, ymin = -Inf, ymax = Inf), fill = "grey95", alpha = 0.5, inherit.aes = FALSE) +
theme_bw()