From Poisson Regression by hand this 'manual' Poisson coefficient function is provided:
LogLike <- function(y,x, par) {
beta <- par
# the deterministic part of the model:
lambda <- exp(beta%*%t(x))
# and here comes the negative log-likelihood of the whole dataset, given the
# model:
LL <- -sum(dpois(y, lambda, log = TRUE))
return(LL)
}
PoisMod<-function(formula, data){
# # definiere Regressionsformel
form <- formula(formula)
#
# # dataFrame wird erzeugt
model <- model.frame(formula, data = data)
#
# # Designmatrix erzeugt
x <- model.matrix(formula,data = data)
#
# # Response Variable erzeugt
y <- model.response(model)
par <- rep(0,ncol(x))
erg <- list(optim(par=par,fn=LogLike,x=x,y=y)$par)
return(erg)
}
PoisMod(breaks~wool+tension, as.data.frame(daten))
glm(breaks~wool+tension, family = "poisson", data = as.data.frame(daten))
Can any one tell me exactly where the link function is computed here? What would this code look like with an identity link function? I have basic understanding from YouTube videos etc, but no one explains the actual computation.
How would this code look like with offset and weights?
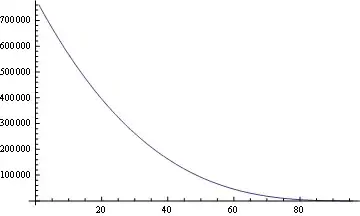 , so
, so