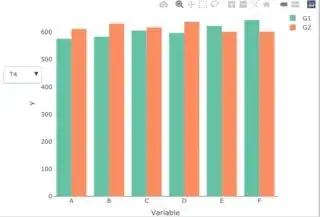
Let's suppose this is my dataset:
set.seed(21022022)
TablaStack <- data.frame(
n <- rbinom(12, 1000, 0.5),
T1 <- rnorm(12, 300, 50),
T2 <- rnorm(12, 500, 60),
T3 <- rnorm(12, 650, 90),
T4 <- rt(12, 500, 600),
Variable <- rep(c("A", "B", "C", "D", "E", "F"), 2),
Group <- c(rep(c("G1"),6),rep(c("G2"),6))
)
colnames(TablaStack) = c("Cases", "T1", "T2", "T3", "T4", "Variable", "Group")
Treatments <- c("Cases", "T1", "T2", "T3", "T4")
My intention is to show the two values (group 1 and 2) of the variables for each treatment with a dropdown menu just like this. This is the code of my visualization, it's an adaptation of this answer:
create_buttons <- function(df, y_axis_var_names) {
lapply(
y_axis_var_names,
FUN = function(var_name, df) {
button <- list(
method = 'restyle',
args = list('y', list(df[, var_name])),
label = sprintf('%s', var_name)
)
},
df
)
}
plot_ly(
TablaStack,
x = ~Variable,
y = ~Cases,
color = ~as.factor(Group),
colors = "Set2",
type = "bar") %>%
layout(
yaxis = list(title = "y"),
updatemenus = list(
list(
y = 0.7,
buttons = create_buttons(TablaStack,
Treatments)
)))
My problem it's when I select a treatment, it doesn't show the values for group 2. I don't understand why this happens.