Ahead of a the programming question, I believe I need to give a little background of what I am doing to ease the understanding of my problem :
I record eye-movements while displaying some patterns to subjects subject. Through the experiment, I later display some symmetrical transform of those patterns.
What I get is lists of fixations coordinates and durations :
{{fix1X,fix1Y,fix1Dn},{fix2X,fix2Y,fix2Dn},...{fixNX,fixNY,fixNDn}}
Where :
-fix1X is the X coordinate for the first fixation.
-fix1Y is the Y coordinate for the first fixation.
-fix1D is the duration in milliseconds of the fixations
Please consider :
FrameWidth = 31.36;
scrHeightCM = 30;
scrWidthCM = 40;
FrameXYs = {{4.32, 3.23}, {35.68, 26.75}}; (* {{Xmin,Ymin},{Xmax,Ymax}} *)
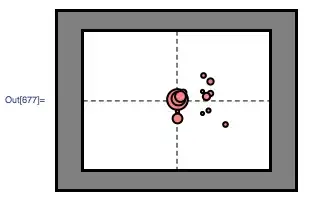
Below are the fixations for 1 Display (Subject Fixations during the 3s stimuli presentation on the screen)
fix ={{20.14, 15.22, 774.}, {20.26, 15.37, 518.}, {25.65, 16.22, 200.},
{28.15, 11.06, 176.}, {25.25, 13.38, 154.}, {24.78, 15.74, 161.},
{24.23, 16.58, 121.}, {20.06, 13.22, 124.}, {24.91, 15.8, 273.},
{24.32, 12.83, 119.}, {20.06, 12.14, 366.}, {25.64, 18.22, 236.},
{24.37, 19.2, 177.}, {21.02, 16.4, 217.}, {20.63, 15.75,406.}}
Graphics[{
Gray, EdgeForm[Thick],
Rectangle @@ {{0, 0}, {scrWidthCM, scrHeightCM}},
White,
Rectangle @@ StimuliFrameCoordinates,
Dashed, Black,
Line[
{{(scrWidthCM/2), FrameXYs[[1, 2]]},
{(scrWidthCM/2), FrameXYs[[2, 2]]}}],
Line[
{{FrameXYs[[1, 1]], (scrHeightCM/2)},
{(FrameXYs[[2, 1]]), (scrHeightCM/2)}}],
Thickness[0.005], Pink,
Disk[{#[[1]], #[[2]]}, 9 N[#[[3]]/Total[fix[[All, 3]]]]] & /@ fix
}, ImageSize -> 500]
What I want to do :
I would like to "discretize" the stimuli frame space into clusters :
Below is a visual representation (done in PPT) with different clusters (2,4,16,64).
The colored part representing the clusters in which fixations occurred :

With this I want to
-Count the number of fixations within each cluster.
-Compute the presence/count or duration observed in each cluster.
A Matrix form would easily enable me to compare different displays fixations by subtraction.
So, question(s)
-How can I create a flexible mechanism to divide the stimuli frame into clusters.
-Map the fixations onto those clusters obtaining a rectangular Matrix filled with 0s or fixations counts or total fixations duration for each of the matrix cell.
I feel this question could be unclear and will edit it to clarify anything needed. Also, would you think this should be asked in 2 separate questions, I am happy to do so.
Many thank in advance for any help you could provide.