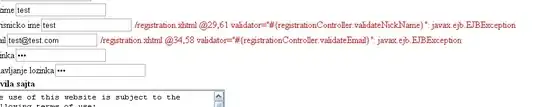
I tried to take the average of SF_Plante_Verte and SF_Plante_Totale acccording to their Date_obs.
df<-structure(list(Pos_heliaphen = c("X47", "W17", "Z17", "X47",
"Y19", "Y40", "X47", "Y19", "Y40", "Z17", "Z31", "X47", "Y19",
"Y40", "Z31", "X47", "Z17", "Z31"), traitement = c("WW", "WW",
"WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW",
"WW", "WW", "WW", "WW", "WW"), Variete = c("Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas"), Date_obs = c("D11_04/06/2021",
"D11_04/06/2021", "D11_04/06/2021", "D12_07/06/2021", "D12_07/06/2021",
"D12_07/06/2021", "D23_25/06/2021", "D23_25/06/2021", "D23_25/06/2021",
"D23_25/06/2021", "D23_25/06/2021", "D24_28/06/2021", "D24_28/06/2021",
"D24_28/06/2021", "D24_28/06/2021", "D25_29/06/2021", "D25_29/06/2021",
"D25_29/06/2021"), SF_Plante_Totale = c(46473, 44589.3, 43134,
166645.5, 119962.5, 93061.5, 483583.8, 313985.7, 273432.6, 414871.8,
426766.2, 539410.2, 337417.5, 273432.6, 474915, 539410.2, 414871.8,
474915), SF_Plante_Verte = c(46473, 44589.3, 43134, 162512.7,
119962.5, 93061.5, 462655.2, 293367.9, 238373.1, 363123.6, 407572.2,
473793.6, 316799.7, 238373.1, 420682.5, 473793.6, 363123.6, 420682.5
), SF_Plante_senescence = c(0, 0, 0, 4132.8, 0, 0, 20928.6, 20617.8,
35059.5, 51748.2, 19194, 65616.6, 20617.8, 35059.5, 54232.5,
65616.6, 51748.2, 54232.5)), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -18L))
With the code below, I want to draw a dotted line, but I want to get smooth curves instead of polylines (no polyline segments).And I can't add legend successfully either.
Could anyone please help save my problem? Thank you in advance!
ggplot(df, aes(x = Date_obs)) +
stat_summary(aes(y = SF_Plante_Totale,group=1), fun =mean, colour="white",shape=21,size=4,fill="steelblue",geom="point",group=1)+
stat_summary(aes(y = SF_Plante_Totale,group=1), fun =mean,colour="steelblue", geom="smooth", group=1)+
stat_summary(aes(y = SF_Plante_Verte,group=1), fun =mean, colour="white",shape=21,size=4,fill="tomato",geom="point",group=2)+
stat_summary(aes(y = SF_Plante_Verte,group=1), fun =mean,colour="tomato", geom="smooth", group=1)+
theme(axis.text.x = element_text(angle = 45, hjust = 1))