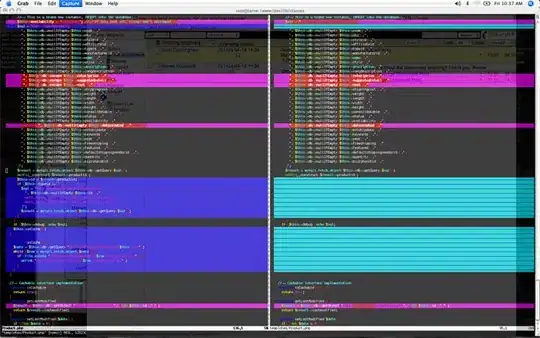
I'm trying to make a stacked bar chart, but the bars overlap.
This is the code that I am using:
library(ggplot2)
ggplot(data = opstaR, aes(x = Indicator, y = Percentage, fill = factor(tip))) +
geom_bar(position = "dodge", stat = "identity") +
coord_flip() +
scale_fill_grey(labels = c("Case load change", "Compliance change")) +
theme_bw() +
scale_y_continuous(labels = scales::percent, limits = c(-0.15, 0.05),breaks = seq(-0.15, 0.05, 0.05)) +
theme(axis.title.y = element_blank()) +
theme(legend.title = element_blank()) +
theme(legend.position = "bottom")
This is the output:
How can I make the bars not stack to each other?
And how to invert the black/gray pattern?
Data
opstaR <- structure(list(Indicator = structure(c(1L, 2L, 3L, 4L, 5L, 6L,
7L, 8L, 9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 18L, 1L,
2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 14L, 15L,
16L, 17L, 18L), .Label = c("HbA1c measurment among diabetic patients",
"LDL-cholesterol measurement among diabetic patients", "Diet advice among diabetic patients",
"Blood preasure measurment among diabetic patients", "Foot examination among diabetic patients",
"Blood preasure among hypertensive patients", "Total cholesterol among hypertensive patients",
"Blood glucose among hypertensive patients", "Lifestyle advice among hypertensive patients",
"ECG among hypertensive patients", "Lifestyle advice among patients with MI",
"Blood preasure measurment among patients with MI", "Total cholesterol among patients with MI",
"LDL-cholesterol measurement among patients with MI", "HDL-cholesterol measurement among patients with MI",
"Triglycerides measurement among patients with MI", "Blood glucose among patients with MI",
"ECG among patients with MI"), class = c("ordered", "factor")),
Percentage = c(-0.119461152, -0.119461152, -0.119461152,
-0.119461152, -0.119461152, -0.125256348, -0.125256348, -0.125256348,
-0.125256348, -0.125256348, -0.139557636, -0.139557636, -0.139557636,
-0.139557636, -0.139557636, -0.139557636, -0.139557636, -0.139557636,
-0.016017788, -0.01085703, 0.030175594, -0.02377841, -0.011164678,
-0.020884472, -0.014592098, -0.013684149, 0.033509796, -0.009671902,
0.037509542, -0.018565706, -0.01177768, -0.007261598, -0.005188794,
-0.008428487, -0.010241197, 0.002690945), tip = c("Change_in_case_load",
"Change_in_case_load", "Change_in_case_load", "Change_in_case_load",
"Change_in_case_load", "Change_in_case_load", "Change_in_case_load",
"Change_in_case_load", "Change_in_case_load", "Change_in_case_load",
"Change_in_case_load", "Change_in_case_load", "Change_in_case_load",
"Change_in_case_load", "Change_in_case_load", "Change_in_case_load",
"Change_in_case_load", "Change_in_case_load", "Change_in_compliance_rate",
"Change_in_case_load", "Change_in_compliance_rate", "Change_in_case_load",
"Change_in_compliance_rate", "Change_in_case_load", "Change_in_compliance_rate",
"Change_in_case_load", "Change_in_compliance_rate", "Change_in_case_load",
"Change_in_compliance_rate", "Change_in_case_load", "Change_in_compliance_rate",
"Change_in_case_load", "Change_in_compliance_rate", "Change_in_case_load",
"Change_in_compliance_rate", "Change_in_case_load")), row.names = c(NA,
-36L), class = "data.frame")