I'm trying to plot a merged plot for bonferroni p adjusted value using ggplot.
I'm running this code
p <- ggplot(stack, aes(x = factor(phenotypes), y = values)) +
geom_boxplot(aes(fill = factor(modules))) +
theme_prism() +
theme(legend.position = "none")
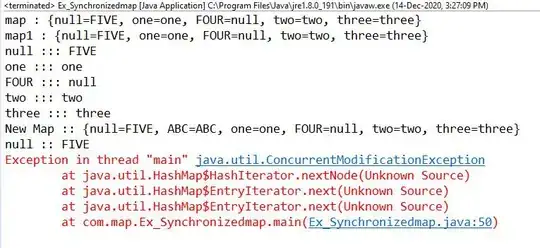
Everytime I run the I get the similar classic error.
Error in grid.Call(C_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
polygon edge not found
The usual problem is font not being presnt in the correct directory. But that doesn't seem to be the issue here.
I'm using this website for reference
EDIT: This is the data, i.e. the "stack" dataset I'm working on.
dput(stack[1:171,])
structure(list(phenotypes = c("Proneural", "Mesenchymal", "Classical",
"Classical", "Proneural", "Proneural", "Proneural", "Classical",
"Classical", "Classical", "Proneural", "Mesenchymal", "Proneural",
"Classical", "Proneural", "Mesenchymal", "Mesenchymal", "Classical",
"Mesenchymal", "Classical", "Proneural", "Proneural", "Proneural",
"Mesenchymal", "Mesenchymal", "Mesenchymal", "Mesenchymal", "Proneural",
"Mesenchymal", "Proneural", "Classical", "Proneural", "Mesenchymal",
"Mesenchymal", "Proneural", "Proneural", "Mesenchymal", "Mesenchymal",
"Proneural", "Neural", "Mesenchymal", "Classical", "Proneural",
"Mesenchymal", "Mesenchymal", "Mesenchymal", "Neural", "Proneural",
"Neural", "Classical", "Classical", "Proneural", "Classical",
"Classical", "Classical", "Classical", "Proneural", "Proneural",
"Mesenchymal", "Classical", "Mesenchymal", "Proneural", "Proneural",
"Classical", "Classical", "Proneural", "Proneural", "Proneural",
"Proneural", "Neural", "Mesenchymal", "Mesenchymal", "Classical",
"Classical", "Proneural", "Proneural", "Mesenchymal", "Neural",
"Neural", "Classical", "Neural", "Mesenchymal", "Mesenchymal",
"Classical", "Proneural", "Mesenchymal", "Classical", "Mesenchymal",
"Mesenchymal", "Mesenchymal", "Proneural", "Neural", "Neural",
"Mesenchymal", "Proneural", "Neural", "Neural", "Neural", "Proneural",
"Mesenchymal", "Mesenchymal", "Proneural", "Neural", "Neural",
"Mesenchymal", "Neural", "Classical", "Mesenchymal", "Mesenchymal",
"Mesenchymal", "Neural", "Mesenchymal", "Neural", "Mesenchymal",
"Classical", "Neural", "Neural", "Neural", "Neural", "Proneural",
"Neural", "Proneural", "Mesenchymal", "Classical", "Mesenchymal",
"Proneural", "Mesenchymal", "Proneural", "Proneural", "Mesenchymal",
"Mesenchymal", "Proneural", "Proneural", "Proneural", "Classical",
"Proneural", "Mesenchymal", "Proneural", "Proneural", "Neural",
"Proneural", "Mesenchymal", "Proneural", "Classical", "Classical",
"Classical", "Classical", "Proneural", "Mesenchymal", "Classical",
"Neural", "Proneural", "Neural", "Mesenchymal", "Mesenchymal",
"Mesenchymal", "Mesenchymal", "Classical", "Mesenchymal", "Classical",
"Proneural", "Classical", "Neural", "Mesenchymal", "Proneural",
"Classical", "Classical", "Proneural", "Proneural", "Mesenchymal",
"Mesenchymal"), values = c(-0.126482404651362, 0.00859685106988051,
-0.139119978025995, -0.121760256216002, -0.114050357589663, -0.16158166178197,
-0.169089521235389, 0.134388350128016, -0.128728040505512, 0.0933639502568886,
-0.116285533417, -0.0715164710720025, -0.050281653395796, -0.0712583935347317,
-0.116848802717176, -0.1394796603133, -0.131958454866075, -0.167862157710569,
0.0781961256653059, -0.0874083890994826, -0.142602528126273,
-0.132749359004561, 0.0530754944359762, -0.112556115187184, -0.126475329197849,
-0.103764754820242, -0.0377897554734426, -0.0825700087419535,
0.0565802594387166, -0.127113477946288, 0.0621406527851034, -0.0353373248499261,
0.0651104404770428, -0.046842885259204, -0.0388244031159389,
-0.0331569959442778, 0.033701438947606, 0.0642070914755685, -0.0753508936454846,
0.116575055033346, 0.0618127428574358, 0.0913128306164853, -0.131098598718593,
0.00663000043165442, 0.087646298412118, 0.0424501375261861, 0.132271826019638,
-0.116098261441119, 0.150578014972223, 0.0370869606412521, 0.124207975834675,
-0.0697137156198783, 0.045603425091438, 0.128723238203833, -0.00899840307387745,
0.0716898207350761, 0.0278474335783047, -0.00251548498551291,
0.00944990607573422, 0.063040030569466, -0.00966936428286957,
-0.103167359695479, -0.109970747377413, 0.092822101720664, 0.0599647788263245,
-0.0402260247392427, 0.0561493322564905, -0.0337741476357838,
0.0123028680258031, 0.0829652405103008, 0.0473861745713383, 0.0552591953919883,
0.0589190391293063, 0.0542800207749991, -0.0590957568660261,
-0.0768057537931389, -0.0631594939938725, 0.0232166721127454,
0.0794818086402467, 0.0163758735536393, 0.0364047314888439, -0.056987419443193,
0.0102418241805152, 0.0834466785300582, -0.0621321025318891,
-0.0202384484936535, 0.0472178852980202, 0.0500099068565177,
-0.000386746443747292, 0.0336169099739554, -0.0196318551702082,
0.0215504671153622, 0.0370043737532354, 0.00236114001435378,
0.0187774063941158, 0.0118748174856794, 0.0438428539755876, 0.0590938337833555,
-0.0214051371142639, 0.048970263838942, -0.0353986999749805,
-0.0652888103586655, 0.0307980036631203, 0.0435024586638615,
0.06349210003803, 0.0699162740441559, 0.0866779470756711, 0.0302570428387929,
0.039907529282083, 0.0413550598629916, 0.0194677506824577, 0.0382215014456658,
0.0998571537054834, 0.0249023251158023, 0.0491338848762949, 0.0739696647120266,
-0.0156915725176812, -0.086181923772576, 0.0484264963060224,
-0.0952872280628109, -0.0496917131983185, -0.0626613426906559,
-0.0235720315114444, 0.0402051758624549, 0.0541716193143272,
-0.0447085228859518, 0.0851145904041888, -0.0750319738540656,
-0.0395468309394087, -0.0171796695631005, 0.0143116629560395,
0.0139348413765643, -0.0605286733223814, -0.0680551202279635,
0.0935675698428226, -0.121323835109815, 0.0650088114184009, 0.0286512581822918,
-0.0267822397048852, 0.0560594126094381, -0.0866917309227976,
0.0761270963342822, 0.0102442673059245, 0.064812961280754, 0.0566119307303998,
0.0903385384522695, -0.0471706176728432, 0.00751391691722696,
0.0350567757887319, 0.0587473888278628, 0.0645068776643195, -0.0843968892218576,
0.0978188183251172, -0.0248768078605493, -0.101713696034819,
0.0535916907441913, 0.081542193680387, 0.107977971088651, 0.076239889269626,
0.0817493803790159, -0.00988348985129774, 0.119297845556547,
0.0769790743428255, -0.0632279011942837, -0.140594814112487,
0.0982145914982851, 0.129445220757845, -0.0740271756065673, -0.049940713932579,
-0.0076347504220374, 0.00346005068662671), modules = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), .Label = c("MEdarkturquoise",
"MEtan", "MEdarkgrey", "MEsalmon", "MEviolet", "MEdarkolivegreen",
"MEdarkred", "MEskyblue", "MEskyblue3", "MEsteelblue", "MEmidnightblue",
"MEcyan", "MEsienna3", "MEyellowgreen", "MElightcyan", "MEorange",
"MEblack", "MEdarkmagenta", "MEdarkorange", "MEwhite", "MEblue",
"MEgrey60", "MEdarkgreen", "MEred", "MEgrey"), class = "factor")), row.names = c(NA,
171L), class = "data.frame")
It has a total of 4275 rows and 3 columns.
Suggestions shall be welcomed.