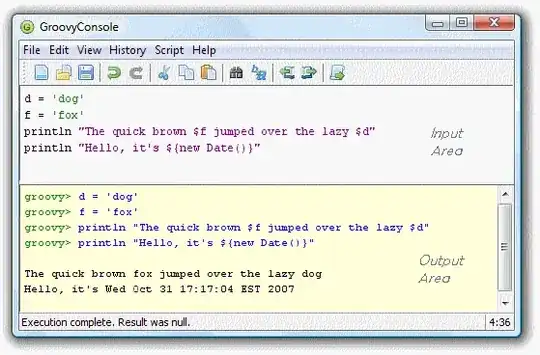
I have this code below that plots the near air surface temperature of greenland, but, I need to mask out the colors outside Greenland and also the islands outside the main island of Greenland.
Below is my code:
import numpy as np
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.mpl.ticker as cmt
import xarray as xr
import matplotlib.ticker as ticker
# Plot func.
def PlotMap(lon,lat,var1,var2,var3,levs, mapbar,unidades, titulo, figura):
parameters = {'xtick.labelsize':14,
'ytick.labelsize':14,
'axes.labelsize':14,
'boxplot.whiskerprops.linewidth':4,
'boxplot.boxprops.linewidth': 4,
'boxplot.capprops.linewidth':4,
'boxplot.medianprops.linewidth':4,
'axes.linewidth':1.5}
plt.rcParams.update(parameters)
fig, ax = plt.subplots(figsize=(9,10))
ax=plt.subplot(1,1,1)
im=ax.contourf(lon,lat,var1,levs, cmap = mapbar)
ax.contour(lon,lat,var2,0,colors='black')
ax.contour(lon,lat,var3,0,colors='red')
#ax.contour(lon,lat,var3,0,colors='blue')
fig.colorbar(im,orientation = 'vertical',shrink=0.8, label = unidades, pad=0.05)
fig.suptitle(titulo, fontsize='large',weight='bold')
plt.tight_layout()
fig.savefig(figur, bbox_inches='tight',pad_inches = 0, dpi=400);
# Open file
grl = xr.open_dataset('/Users/jacobgarcia/Desktop/Master en Meteorologia/TFM/Trabajo fin de master/OUTPUTS/output/ens1/0/yelmo2d.nc')
topo = xr.open_dataset('/Users/jacobgarcia/Desktop/Master en Meteorologia/TFM/Trabajo fin de master/Greenland/GRL-16KM/GRL-16KM_TOPO-M17.nc')
xc = grl['xc'].data
yc = grl['yc'].data
smb = grl['T_srf'].mean(dim="time")
border = grl['z_srf'].mean(dim="time")
border_mask = grl['z_srf'].mean(dim="time").data
region_mask = topo['mask'].data
new_smb = smb.where(region_mask>1,np.nan)
new_border = border.where(border_mask<1290,np.nan)
#bord = grl['regions']
#bord_mask = grl['regions'].data
#new_border2 = bord.where(bord_mask>1.3,np.nan)
#cmin=np.min(zbed)
#cmax=np.max(zbed)+10
cmin=np.min(smb)
cmax= np.max(smb) + 10
levels=np.arange(cmin,cmax,10)
mapbar='bwr'
titulo='Greenland Temp'
unidades='K'
figur='/Users/jacobgarcia/Desktop/Master en Meteorologia/TFM/figs/smb.png'
PlotMap(xc,yc,smb,new_border,smb,levels,mapbar,unidades, titulo, figur)
The data is stored (https://drive.google.com/drive/folders/10XvLZPC9vX9odkfU6aV6qBIxRe9IQvyZ?usp=sharing)
This is a picture of the output of my code:
Any help will be much appreciated.