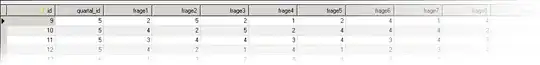
For the dataset df (the data is posted in the end of this question), I try to use gg_season() and gghighlight(year >= 2020) to highlight the lines after the years of 2020.
But the output is obviously not as expected, how could I modify to year >= 2020 to achieve the correct effect of plot? Thanks.
library(tidyverse)
library(lubridate)
library(feasts)
library(tsibble)
library(gghighlight)
df %>%
mutate(date = as.Date(date, origin = "1899-12-30")) %>%
mutate(year=year(date)) %>%
pivot_longer(`food_index`:`energy_index`) %>%
mutate(date=yearmonth(date)) %>%
as_tsibble(index=date, key=name) %>%
gg_season(value, alpha=1) +
geom_line(size=0.8, alpha=0.8) +
geom_point(size=2, alpha=1) +
gghighlight(year >= 2020)
Out:
Data:
df <- structure(list(date = c(42766, 42794, 42825, 42855, 42886, 42916,
42947, 42978, 43008, 43039, 43069, 43100, 43131, 43159, 43190,
43220, 43251, 43281, 43312, 43343, 43373, 43404, 43434, 43465,
43496, 43524, 43555, 43585, 43616, 43646, 43677, 43708, 43738,
43769, 43799, 43830, 43861, 43890, 43921, 43951, 43982, 44012,
44043, 44074, 44104, 44135, 44165, 44196, 44227, 44255, 44286,
44316, 44347, 44377, 44408, 44439, 44469, 44500, 44530, 44561
), food_index = c(58.53, 61.23, 55.32, 55.34, 61.73, 56.91, 54.27,
59.08, 60.11, 66.01, 60.11, 63.41, 69.8, 72.45, 81.11, 89.64,
88.64, 88.62, 98.27, 111.11, 129.39, 140.14, 143.44, 169.21,
177.39, 163.88, 135.07, 151.28, 172.81, 143.82, 162.13, 172.22,
176.67, 179.3, 157.27, 169.12, 192.51, 194.2, 179.4, 169.1, 193.17,
174.92, 181.92, 188.41, 192.14, 203.41, 194.19, 174.3, 174.86,
182.33, 182.82, 185.36, 192.41, 195.59, 202.6, 201.51, 225.01,
243.78, 270.67, 304.57), energy_index = c(127.36, 119.87, 120.96,
112.09, 112.19, 109.24, 109.56, 106.89, 109.35, 108.35, 112.39,
117.77, 119.52, 122.24, 120.91, 125.41, 129.72, 135.25, 139.33,
148.6, 169.62, 184.23, 204.38, 198.55, 189.29, 202.47, 220.23,
240.67, 263.12, 249.74, 240.84, 243.42, 261.2, 256.76, 258.69,
277.98, 289.63, 293.46, 310.81, 318.68, 310.04, 302.17, 298.62,
260.92, 269.29, 258.84, 241.68, 224.18, 216.36, 226.57, 235.98,
253.86, 267.37, 261.99, 273.37, 280.91, 291.84, 297.88, 292.78,
289.79)), row.names = c(NA, 60L), class = "data.frame")