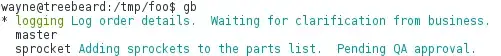
This what I have:
x = df2['bulk_TPM']
y = df2['scRNA_TPM']
plt.scatter(x,y,cmap ='summer',
edgecolor='black',linewidth=1, alpha = 0.75)
plt.xscale('log')
plt.yscale('log')
plt.title('bulk vs scRNA log10 TPM')
plt.xlabel('bulk_TPM')
plt.ylabel('scRNA_TPM')
I attempted changing the scale by doing this:
plt.xscale('log').xlim(0,10**5)
and same method for y-axis but get weird scale size.
I would like both x and y axis to have same 0-10^5, how can I resolve this