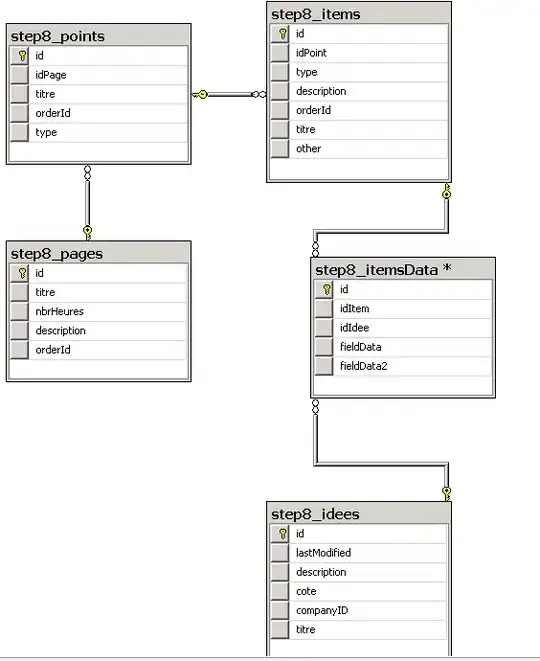
This can be done using Pillow (fork of PIL) as follows:
from PIL import Image
import numpy as np
n = 3 # repeatation
im = Image.fromarray(arr)
up_im = im.resize((im.width*n, im.height*n),resample=Image.NEAREST)
up_arr = np.array(up_im)
Example:
arr = np.array(
[[0, 0, 0, 1, 1],
[0, 1, 1, 1, 1],
[1, 1, 0, 0, 1],
[0, 0, 1, 0, 1],
[0, 1, 1, 0, 1]])
res (n=3):
np.array(
[[0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1],
[0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1],
[0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1],
[0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1],
[1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1],
[1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1],
[1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1],
[0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 1],
[0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 1],
[0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 1],
[0, 0, 0, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1, 1, 1],
[0, 0, 0, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1, 1, 1],
[0, 0, 0, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1, 1, 1]])
numba is by far the most performant in terms of speed. As the matrix size increases, PIL takes much more time.