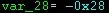Given this GPS dataset (sample.csv) from Beijing, I am trying to apply pyKalman so as to fill the gaps on the GPS series. The code I am using is taken from here:
from pykalman import KalmanFilter
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
def main():
method = 1
coords = pd.read_csv('sample.csv')
# coords.index = pd.to_datetime(coords.index)
coords['time'] = pd.to_datetime(coords['time'])
coords = coords.sort_values(by="time").reset_index(drop=True)
# coords = coords[coords['time'].ne(coords['time'].shift())]
coords.set_index('time', inplace=True, drop=True)
coords = coords[~coords.index.duplicated()]
coords['idx'] = range(0, len(coords))
if method == 1:
print(coords.head())
coords.index = np.round(coords.index.astype(np.int64), -9).astype('datetime64[ns]')
coords = coords.resample('1S').asfreq()
measurements = np.ma.masked_invalid(coords[['lon', 'lat', 'alt']].values)
else:
import datetime
METHOD_MAX_GAP = 20 # seconds
"""
Method 2: fill the gaps between points close to each other's with NaNs and leave the big holes alone.
The resulting sampling time is not constant
"""
for i in range(0, len(coords) - 1):
gap = coords.index[i + 1] - coords.index[i]
if datetime.timedelta(seconds=1) < gap <= datetime.timedelta(seconds=METHOD_MAX_GAP):
gap_idx = pd.date_range(start=coords.index[i] + datetime.timedelta(seconds=1),
end=coords.index[i + 1] - datetime.timedelta(seconds=1),
freq='1S')
gap_coords = pd.DataFrame(coords, index=gap_idx)
coords = coords.append(gap_coords)
# print("Added {} points in between {} and {}"
# .format(len(gap_idx), coords.index[i], coords.index[i + 1]))
# Sort all points
coords = coords.sort_index()
# Fill the time_sec column
coords['time_sec'] = np.nan
for i in range(0, len(coords)):
coords['time_sec'].values[i] = (coords.index[i] - datetime.datetime(2000, 1, 1, 0, 0, 0)).total_seconds()
coords['time_sec'] = coords['time_sec'] - coords['time_sec'][0]
# Create the "measurement" array and mask NaNs
measurements = coords[['lon', 'lat', 'alt']].values
measurements = np.ma.masked_invalid(measurements)
# Covariances: Position = 0.0001deg = 11.1m, Altitude = 30m
cov = {'coordinates': 1.,
'elevation': 30.,
'horizontal_velocity': 1e-3,
'elevation_velocity': 1e-3,
'horizontal_acceleration': 1e-6 * 1000,
'elevation_acceleration': 1e-6 * 1000}
if method == 1:
transition_matrices = np.array([[1, 0, 0, 1, 0, 0],
[0, 1, 0, 0, 1, 0],
[0, 0, 1, 0, 0, 1],
[0, 0, 0, 1, 0, 0],
[0, 0, 0, 0, 1, 0],
[0, 0, 0, 0, 0, 1]])
else:
# The samples are randomly spaced in time, so dt varies with time and a
# time dependent transition matrix is necessary
timesteps = np.asarray(coords['time_sec'][1:]) - np.asarray(coords['time_sec'][0:-1])
transition_matrices = np.zeros(shape=(len(timesteps), 6, 6))
for i in range(len(timesteps)):
transition_matrices[i] = np.array([[1., 0., 0., timesteps[i], 0., 0.],
[0., 1., 0., 0., timesteps[i], 0.],
[0., 0., 1., 0., 0., timesteps[i]],
[0., 0., 0., 1., 0., 0.],
[0., 0., 0., 0., 1., 0.],
[0., 0., 0., 0., 0., 1.]])
observation_matrices = np.array([[1, 0, 0, 0, 0, 0],
[0, 1, 0, 0, 0, 0],
[0, 0, 1, 0, 0, 0]])
# observation_covariance = np.diag([1e-4, 1e-4, 100]) ** 2
observation_covariance = np.diag([cov['coordinates'], cov['coordinates'], cov['elevation']]) ** 2
initial_state_mean = np.hstack([measurements[0, :], 3 * [0.]])
# works initial_state_covariance = np.diag([1e-3, 1e-3, 100, 1e-4, 1e-4, 1e-4])**2
# initial_state_covariance = np.diag([1e-4, 1e-4, 50, 1e-6, 1e-6, 1e-6]) ** 2
initial_state_covariance = np.diag([cov['coordinates'], cov['coordinates'], cov['elevation'],
cov['horizontal_velocity'], cov['horizontal_velocity'],
cov['elevation_velocity']]) ** 2
kf = KalmanFilter(transition_matrices=transition_matrices,
observation_matrices=observation_matrices,
observation_covariance=observation_covariance,
initial_state_mean=initial_state_mean,
initial_state_covariance=initial_state_covariance,
# em_vars=['transition_matrices',
# 'observation_matrices',
# 'transition_offsets',
# 'observation_offsets',
# 'transition_covariance',
# 'observation_covariance',
# 'initial_state_mean',
# 'initial_state_covariance'
# ]
)
kf = kf.em(measurements, n_iter=5)
state_means, state_vars = kf.smooth(measurements)
# Increase observation co-variance
kf.observation_covariance = kf.observation_covariance * 10
filled_coords = state_means[coords['idx'].isnull(), :3]
orig_coords = coords[~coords['idx'].isnull()].set_index('idx')
plt.plot(orig_coords['lon'], orig_coords['lat'], 'r.', markersize=2, label='original')
plt.plot(filled_coords[:, 0], filled_coords[:, 1], 'go', markersize=2, label='filled')
plt.legend()
plt.show()
if __name__ == '__main__':
main()
The output of method 1 is
While the output of method 2 is  . Could you please explain why
. Could you please explain why method 1 returns this strange result? In addition, in method 2, the upper right part of the GPS series is not filled. Is that because of wrong initialization?