Since we don’t have access to your data I simulate fake data in a square.
There are several options to do what you want. First you should know that
density() is a generic function, so when you invoke it on a ppp like
Microglia_PPP actually the function density.ppp() is invoked.
This function returns an im object (effectively a 2-d “image” of values).
You plot this with plot() which in turn calls plot.im(), so you should
read the help file of plot.im(), where it says that the argument col
controls the colours used in the plot. Either you can make a colour map
covering the range of values you are interested in and supply that, or if you
know that one of the images has the colour map you want to use you can save
it and reuse for the others:
library(spatstat)
set.seed(42)
Microglia_PPP <- runifpoint(100)
Neurons_PPP <- runifpoint(200)
Neurons_Density <- density(Neurons_PPP, sigma = 0.1)
Microglia_Density <- density(Microglia_PPP, sigma = 0.1)
my_colourmap <- plot(Neurons_Density, main = "Neuronal density", col = topo.colors)
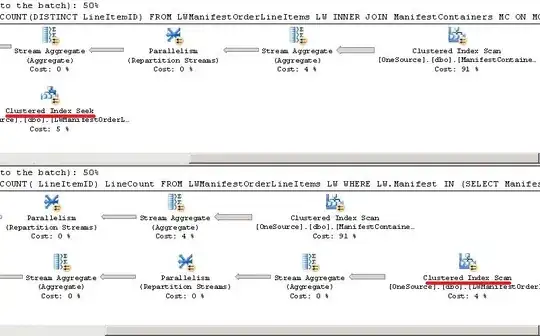
plot(Microglia_Density, main = "Microglia density", col = my_colourmap)
Notice the colour maps are the same, but it only covers the range from
approximately 80 to 310. Any values of the image outside this range will not
be plottet, so they appear white.
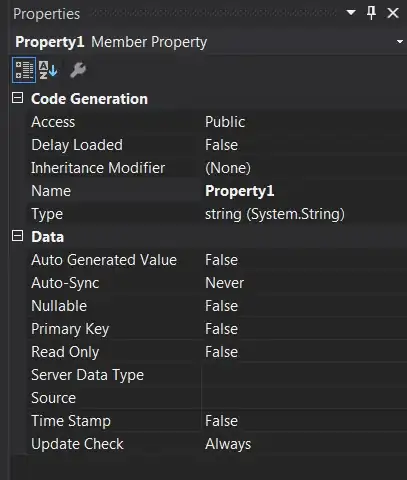
You can make a colour map first and then use it for all the plots
(see help(colourmap)):
my_colourmap <- colourmap(topo.colors(256), range = c(40,315))
plot(Neurons_Density, main = "Neuronal density", col = my_colourmap)
plot(Microglia_Density, main = "Microglia density", col = my_colourmap)
Finally another solution if you want the images side by side is to make them
an imlist (image list) and use plot.imlist() with equal.ribbon = TRUE:
density_list <- as.imlist(list(Neurons_Density, Microglia_Density))
plot(density_list, equal.ribbon = TRUE, main = "")