I asked participants questions concerning their health status and they could either choose "yes" or "no". Now, I want to create a subset of participants that reported having no symptoms at all, i.e., only said "no" to EVERY symptom.
So, all in all, I am searching for "no"-entries to create a subset (in R) and examine the number of people that have no symptoms at all.
The thing is that I only assessed headache in Week 1, sickness and fatigue in Week 2, and Coughing and Diarrhea in Week 3. Therefore, I get NA for the missing values.
So far, so good. When I am searching for "yes" to create a subset with participants that reported to have at least ONE symptom (or more), my results are fine. But when I try to do it vice versa, it does not really work, since I just want to have the "no" answers. As soon as, a participant has a symptom I want them to be excluded.
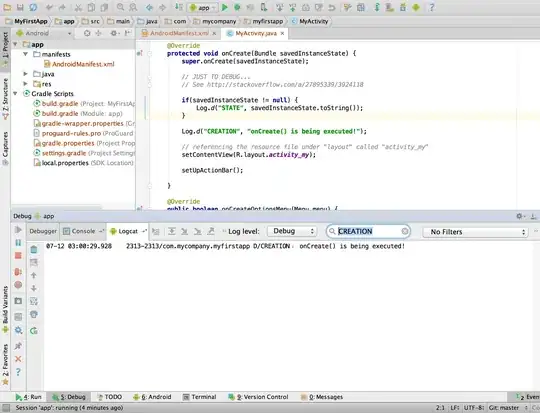
This is what my code looks like:
data$no_symptoms <- case_when(
data$headache == "no" ~ "NS",
data$sickness == "no" ~ "NS",
data$coughing == "no" ~ "NS",
data$fatigue == "no" ~ "NS",
data$diarrhea == "no" ~ "NS",
TRUE ~ as.character(data$headache, data$sickness, data$coughing, data$fatigue,
data$diarrhea)
)
no_symptoms <- subset(data,data$no_symptoms=="NS")
I expected a subset that would look like this if I open it:
I am super grateful for every hint or advice!!! Let me know if I can add some more information. M< main problem is just that I want to exclude everybody that says "yes" across all symptoms. I don't care about NA, I just need the people that said "no" to every assessed symptom.
Thank you so much! :)
Gertie