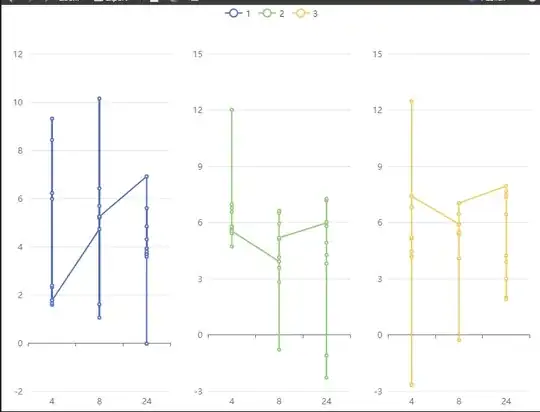
I really like the possibilities this package offers and would like to use it in a shiny app. however i am struggling to recreate a plot from ggplot to echarts4r
library(tidyverse)
library(echarts4r)
data = tibble(time = factor(sort(rep(c(4,8,24), 30)), levels = c(4,8,24)),
dose = factor(rep(c(1,2,3), 30), levels = c(1,2,3)),
id = rep(sort(rep(LETTERS[1:10], 3)),3),
y = rnorm(n = 90, mean = 5, sd = 3))
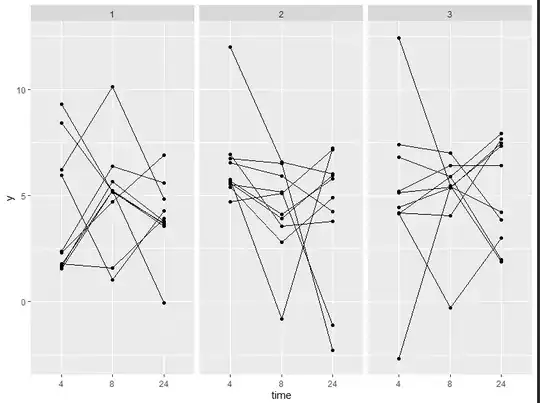
This is the plot i am aiming to recreate:
ggplot(data = data, mapping = aes(x = time, y = y, group = id)) +
geom_point() +
geom_line() +
facet_wrap(~dose)
The problem i am having is to make groups of my data using group = id in ggplot syntax in echarts4r . I am aiming to do e_facet on grouped data using group_by() however i can not (or dont know how to) add a group to connect the dots using geom_line()
data %>%
group_by(dose) %>%
e_charts(time) %>%
e_line(y) %>%
e_facet(rows = 1, cols = 3)