I have a continuous time variable and I want to plot a heatmap of values. To do so using ggplot and geom_tile() I bin the time into time_bin and parse the bounds of the resulting intervals. My question is somewhat related to this one, which has no answer.
Here's a toy data.frame
grouped_df [36 × 5] (S3: grouped_df/tbl_df/tbl/data.frame)
$ y : chr [1:36] "1" "1" "1" "1" ...
$ time_bin: Factor w/ 100 levels "(-0.03,0.3]",..: 1 2 3 4 1 2 3 4 1 2 ...
$ value : num [1:36] -0.4 -0.512 -0.608 -0.725 0.757 ...
$ low : num [1:36] -0.03 0.3 0.601 0.901 -0.03 0.3 0.601 0.901 -0.03 0.3 ...
$ high : num [1:36] 0.3 0.601 0.901 1.2 0.3 0.601 0.901 1.2 0.3 0.601 ...
- attr(*, "groups")= tibble [9 × 2] (S3: tbl_df/tbl/data.frame)
..$ y : chr [1:9] "1" "2" "3" "4" ...
..$ .rows: list<int> [1:9]
.. ..$ : int [1:4] 1 2 3 4
.. ..$ : int [1:4] 5 6 7 8
.. ..$ : int [1:4] 9 10 11 12
.. ..$ : int [1:4] 13 14 15 16
.. ..$ : int [1:4] 17 18 19 20
.. ..$ : int [1:4] 21 22 23 24
.. ..$ : int [1:4] 25 26 27 28
.. ..$ : int [1:4] 29 30 31 32
.. ..$ : int [1:4] 33 34 35 36
.. ..@ ptype: int(0)
..- attr(*, ".drop")= logi TRUE
> dput(toy)
structure(list(y = c("1", "1", "1", "1", "2", "2", "2", "2",
"3", "3", "3", "3", "4", "4", "4", "4", "5", "5", "5", "5", "6",
"6", "6", "6", "7", "7", "7", "7", "8", "8", "8", "8", "9", "9",
"9", "9"), time_bin = structure(c(1L, 2L, 3L, 4L, 1L, 2L, 3L,
4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L,
4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L), .Label = c("(-0.03,0.3]",
"(0.3,0.601]", "(0.601,0.901]", "(0.901,1.2]", "(1.2,1.5]", "(1.5,1.8]",
"(1.8,2.1]", "(2.1,2.4]", "(2.4,2.7]", "(2.7,3]", "(3,3.3]",
"(3.3,3.6]", "(3.6,3.9]", "(3.9,4.2]", "(4.2,4.5]", "(4.5,4.81]",
"(4.81,5.11]", "(5.11,5.41]", "(5.41,5.71]", "(5.71,6.01]", "(6.01,6.31]",
"(6.31,6.61]", "(6.61,6.91]", "(6.91,7.21]", "(7.21,7.51]", "(7.51,7.81]",
"(7.81,8.11]", "(8.11,8.41]", "(8.41,8.71]", "(8.71,9.01]", "(9.01,9.31]",
"(9.31,9.61]", "(9.61,9.91]", "(9.91,10.2]", "(10.2,10.5]", "(10.5,10.8]",
"(10.8,11.1]", "(11.1,11.4]", "(11.4,11.7]", "(11.7,12]", "(12,12.3]",
"(12.3,12.6]", "(12.6,12.9]", "(12.9,13.2]", "(13.2,13.5]", "(13.5,13.8]",
"(13.8,14.1]", "(14.1,14.4]", "(14.4,14.7]", "(14.7,15]", "(15,15.3]",
"(15.3,15.6]", "(15.6,15.9]", "(15.9,16.2]", "(16.2,16.5]", "(16.5,16.8]",
"(16.8,17.1]", "(17.1,17.4]", "(17.4,17.7]", "(17.7,18]", "(18,18.3]",
"(18.3,18.6]", "(18.6,18.9]", "(18.9,19.2]", "(19.2,19.5]", "(19.5,19.8]",
"(19.8,20.1]", "(20.1,20.4]", "(20.4,20.7]", "(20.7,21]", "(21,21.3]",
"(21.3,21.6]", "(21.6,21.9]", "(21.9,22.2]", "(22.2,22.5]", "(22.5,22.8]",
"(22.8,23.1]", "(23.1,23.4]", "(23.4,23.7]", "(23.7,24]", "(24,24.3]",
"(24.3,24.6]", "(24.6,24.9]", "(24.9,25.2]", "(25.2,25.5]", "(25.5,25.8]",
"(25.8,26.1]", "(26.1,26.4]", "(26.4,26.7]", "(26.7,27]", "(27,27.3]",
"(27.3,27.6]", "(27.6,27.9]", "(27.9,28.2]", "(28.2,28.5]", "(28.5,28.8]",
"(28.8,29.1]", "(29.1,29.4]", "(29.4,29.7]", "(29.7,30.1]"), class = "factor"),
value = c(-0.400237814865178, -0.511748154576217, -0.608275784372335,
-0.7247244523613, 0.757330374272958, 0.776718248240655, 0.790724987842227,
0.998768292393676, 1.77979507741577, 1.52945992260953, 1.23688248753942,
0.908341101522855, -1.19238496743088, -1.26961869177189,
-1.32516082947541, -1.38508403291503, -0.629615898978496,
-0.620941659841269, -0.654461627603879, -0.751893145646679,
2.37041367819975, 2.51316431734167, 2.79123079147629, 3.1167959195256,
-0.519653805023003, -0.660372294607146, -0.972330338954077,
-1.31768889807071, 2.11290890332694, 2.21939390456188, 2.21971276073297,
2.15853062905871, 0.0602363209937846, 0.296034218681414,
0.372831656177899, 0.273945402834637), low = c(-0.03, 0.3,
0.601, 0.901, -0.03, 0.3, 0.601, 0.901, -0.03, 0.3, 0.601,
0.901, -0.03, 0.3, 0.601, 0.901, -0.03, 0.3, 0.601, 0.901,
-0.03, 0.3, 0.601, 0.901, -0.03, 0.3, 0.601, 0.901, -0.03,
0.3, 0.601, 0.901, -0.03, 0.3, 0.601, 0.901), high = c(0.3,
0.601, 0.901, 1.2, 0.3, 0.601, 0.901, 1.2, 0.3, 0.601, 0.901,
1.2, 0.3, 0.601, 0.901, 1.2, 0.3, 0.601, 0.901, 1.2, 0.3,
0.601, 0.901, 1.2, 0.3, 0.601, 0.901, 1.2, 0.3, 0.601, 0.901,
1.2, 0.3, 0.601, 0.901, 1.2)), class = c("grouped_df", "tbl_df",
"tbl", "data.frame"), row.names = c(NA, -36L), groups = structure(list(
y = c("1", "2", "3", "4", "5", "6", "7", "8", "9"), .rows = structure(list(
1:4, 5:8, 9:12, 13:16, 17:20, 21:24, 25:28, 29:32, 33:36), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -9L), class = c("tbl_df",
"tbl", "data.frame"), .drop = TRUE))
To plot it, I use
toy %>%
ggplot() +
geom_tile(aes(x=time_bin, y=y, fill=value), color=NA)+
geom_vline(aes(xintercept = unique(time_bin[length(time_bin)/2])))
Which produces
I would like the x axis to be labeled only using the first portion of the breaks (-0.03, 0.3, ...). I have been trying to use scale_x_discrete(breaks, labels) with different approaches but haven't made much progress. The x scale is long in the real data, so ideally it I would be able to do something like scale_x_discrete(breaks=scales::pretty_breaks(5)), but that's also not working.
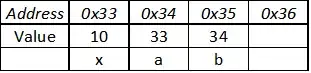
I also tried using the low and high values that I parse from the cut, but that creates tile plots that contain vertical white lines everywhere.
Update
Using insight from one of the answers, I used factor(low) because my parsing works well, but the proposed readr::parse_number() does not. This gets the labels into the proper format.
The remaining portion of the question would be, how to show less factor levels using scale_x_discrete(breaks = ...) ?
For example, this seems to decimate the axis, a bit cumbersome but kinda works (from here)
scale_x_discrete(breaks = function(x){x[c(rep(FALSE, 9), TRUE)]})+
EDIT
For completeness....I am parsing the results from cut using
parse_cuts <- function(x){
# This f(x) is made to be used as
# mutate(map_df(time_bin, function(.x) parse_cuts(as.character(.x))))
out <- sapply(strsplit(x, "\\(|,|]"), function(qq) as.numeric(qq[-1]))[,1]
names(out) <- c("low", "high")
return(unlist(out))
}