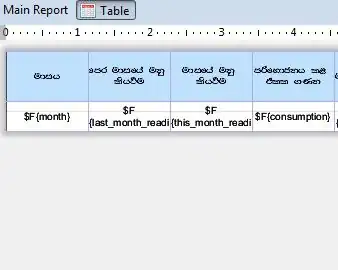
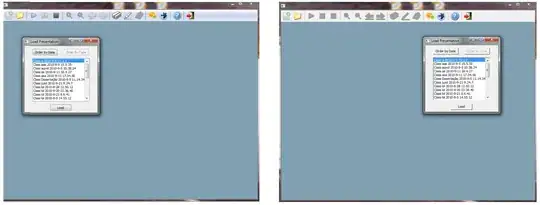
I am trying to plot temperature over time (in the form of a date), however, I am not sure how to.
See here my original table in Excel:
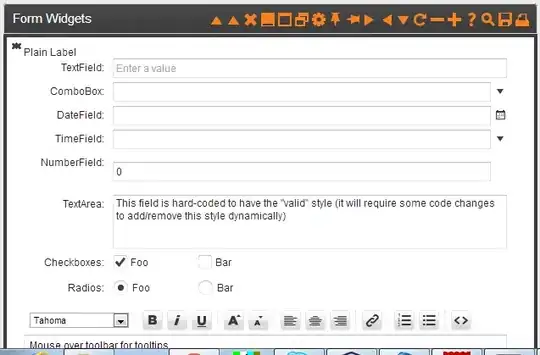
Or as R code:
dput(Average_temperature_period)
structure(list(Sample = c("ZS_IG_1", "AK_SN_1", "JP_IG_2", "AW_IG_1",
"SBB_SN_1", "AW_IG_2", "JvH_IG_3", "JvH_IG_2", "SBB_SN_4", "SBB_SN_3",
"SBB_SN_2", "EF_SN_1", "JP_IG_2", "JvH_IG_3", "EF_SN_1", "JvH_IG_2",
"AK_SN_1", "ZS_IG_1", "AW_IG_1", "SBB_SN_1", "AW_IG_2", "SBB_SN_4",
"SBB_SN_3", "SBB_SN_2"), Sampling_date = c("23/03/2022", "24/03/2022",
"25/03/2022", "25/03/2022", "25/03/2022", "25/03/2022", "29/03/2022",
"29/03/2022", "01/04/2022", "01/04/2022", "01/04/2022", "12/04/2022",
"25/04/2022", "26/04/2022", "28/04/2022", "29/04/2022", "03/05/2022",
"04/05/2022", "10/05/2022", "10/05/2022", "11/05/2022", "11/05/2022",
"12/05/2022", "12/05/2022"), Period = c("March", "March", "March",
"March", "March", "March", "March", "March", "March", "March",
"March", "March", "AprilMay", "AprilMay", "AprilMay", "AprilMay",
"AprilMay", "AprilMay", "AprilMay", "AprilMay", "AprilMay", "AprilMay",
"AprilMay", "AprilMay"), Average_temperature_field = c(7.137037037,
6.966666667, 10.55555556, 7.281481481, 6.874074074, 9.211111111,
9.662962963, 8.12962963, 6.707407407, 6.774074074, 7.162962963,
8.114814815, NA, 11.74814815, 13.51111111, 11.29259259, 15.4962963,
NA, 15.45925926, 17.14814815, 17.72592593, 15.84074074, 16.85555556,
19.78148148), Average_moisture_field = c(33.48518519, 47.35555556,
32.54814815, 34.01851852, 38.66666667, 31.71851852, 23.54814815,
26.83333333, 42.47777778, 29.45555556, 44.50740741, 40.27407407,
25.77407407, 18.91481481, 26.67777778, 16.27407407, 25.38518519,
19.9962963, 18.27777778, 16.14074074, 22.86666667, 23.48518519,
13.93703704, 20.92222222)), row.names = c(NA, 24L), class = "data.frame")
See here my code in R thus far:
##### Soil temperature graph
Average_temperature_period <- read.csv("~/Desktop/First Internship/MicroResp/R/R script/Average_temperature_period.csv")
Average_temperature_period$Sampling_date <- as.character(Average_temperature_period$Sampling_date)
Average_temperature_period <- Average_temperature_period[c(1:24),c(1:5)]
# Change order x axis (past to present)
Average_temperature_period$Sampling_date <- factor(Average_temperature_period$Sampling_date, levels = c("23/03/22","24/03/22","25/03/22","29/03/22","01/04/22","12/04/22","25/04/22","26/04/22","28/04/22","29/04/22","03/05/22","04/05/22","10/05/22","11/05/22","12/05/22"))
# Plot average temperature against the date
ggplot(data=Average_temperature_period, aes(x=Sampling_date, y=Average_temperature_field)) +
geom_smooth(method = "lm", se=FALSE, color="black", aes(group=1)) +
theme_classic() +
ylab("Average soil temperature (°C)") +
xlab("Sampling date")
The x axis keeps on showing 'NA' for the sampling date. Does anyone know why and how to fix it? I would like to have the x axis in order of date (past to present).