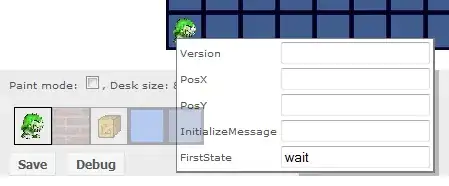
This question is a follow up question to Overlay of forest plot from ZINB model. I need help in displaying the estimated value in the forest plot from ZINB model indicated in the picture below
library(sjPlot)
library(sjlabelled)
library(sjmisc)
library(ggplot2)
library(MASS)
library(pscl)
library(boot)
library(broom)
library(poissonreg)
library(tidyverse) ## purrr::map_dfr, ggplot ...
theme_set(theme_bw())
library(colorspace)
zinb_all_uni <- zeroinfl(ivdays~age,
link="logit",
dist = "negbin",
data=caterpillor)
summary(zinb_all_uni)
plot_model(zinb_all_uni, type="est")
zinb_full_adj <- zeroinfl(ivdays~age+sex+edu,
link="logit",
dist = "negbin",
data=caterpillor)
summary(zinb_full_adj)
plot_model(zinb_full_adj, type="est", terms = c("count_ageb", "count_agec", "zero_ageb", "zero_agec"))
############ second model#######
Zinb_uni_sub <- zeroinfl(ivdays~age,
link="logit",
dist = "negbin",
data=subset(caterpillor, country=="eng"))
summary(Zinb_uni_sub)
plot_model(Zinb_uni_sub, type="est")
zinb_adj_sub <- zeroinfl(ivdays~age+sex+edu,
link="logit",
dist = "negbin",
data=subset(caterpillor, country=="eng"))
summary(zinb_adj_sub)
plot_model(zinb_adj_sub, type="est", terms = c("count_ageb", "count_agec", "zero_ageb", "zero_agec"))
mod_list <- list(all_uni = zinb_all_uni, uni_sub = Zinb_uni_sub)
tidy(zinb_all_uni, type = "all")
coefs <- (mod_list
|> map_dfr(tidy, type = "all",
.id = "model")
## construct CIs
|> mutate(conf.low = qnorm(0.025, estimate, std.error),
conf.high = qnorm(0.975, estimate, std.error))
|> filter(term != "(Intercept)") ## usually don't want this
## cosmetic (strip results down to the components we actually need)
|> dplyr::select(model, term, type, estimate, conf.low, conf.high)
## back-transform
|> mutate(across(c(estimate, conf.low, conf.high), exp))
|> filter(stringr::str_detect(term, "^age"))
)
ggplot(coefs, aes(x = estimate, y = term, colour = model)) +
geom_pointrange(aes(xmin = conf.low, xmax = conf.high),
position = position_dodge(width = 0.5)) +
## separate count-ratio and odds-ratio (conditional/zero) plots
facet_wrap(~type, scale = "free") +
scale_color_discrete_qualitative()+ ## cosmetic
geom_vline(xintercept = 1, lty = 1, color = "yellow", size = 1.5)+
labs(x = "Incidence rate ratio: #IV days", y= "age")
Here is the plots. I would like to display the estimated values circled below:
The following is the data:
caterpillor=structure(list(id = 1:100,
age = structure(c(1L, 1L, 2L, 1L,
2L, 2L, 2L, 3L, 3L, 3L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 3L, 3L, 3L,
1L, 1L, 2L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 1L, 1L, 2L, 1L, 2L, 2L,
2L, 3L, 3L, 3L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 1L, 1L,
2L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 3L,
3L, 3L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 1L, 1L, 2L, 1L,
2L, 2L, 2L, 3L, 3L, 3L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 3L, 3L, 3L),
.Label = c("a", "b", "c"), class = "factor"),
sex = structure(c(2L,
1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L,
1L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L,
2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L,
2L, 1L, 1L),
.Label = c("F", "M"), class = "factor"),
country = structure(c(1L,
1L, 1L, 1L, 3L, 3L, 3L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 3L, 3L, 3L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 2L, 2L, 2L, 1L, 1L, 1L,
1L, 3L, 3L, 3L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 3L,
3L, 3L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 3L, 3L, 3L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 3L, 3L, 3L,
2L, 2L, 2L),
.Label = c("eng", "scot", "wale"), class = "factor"),
edu = structure(c(1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 3L,
1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 2L, 2L,
2L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 3L,
1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 2L, 2L,
2L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 3L,
1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 2L, 2L,
2L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L, 3L),
.Label = c("x", "y", "z"), class = "factor"),
lungfunction = c(45L,
23L, 25L, 45L, 70L, 69L, 90L, 50L, 62L, 45L, 23L, 25L, 45L,
70L, 69L, 90L, 50L, 62L, 45L, 23L, 25L, 45L, 70L, 69L, 90L,
50L, 62L, 45L, 23L, 25L, 45L, 70L, 69L, 90L, 50L, 62L, 45L,
23L, 25L, 45L, 70L, 69L, 90L, 50L, 62L, 45L, 23L, 25L, 45L,
70L, 69L, 90L, 50L, 62L, 45L, 23L, 25L, 45L, 70L, 69L, 90L,
50L, 62L, 45L, 23L, 25L, 45L, 70L, 69L, 90L, 50L, 62L, 45L,
23L, 25L, 45L, 70L, 69L, 90L, 50L, 62L, 25L, 45L, 70L, 69L,
90L, 50L, 62L, 25L, 45L, 70L, 69L, 90L, 50L, 62L, 25L, 45L,
70L, 69L, 90L),
ivdays = c(15L, 26L, 36L, 34L, 2L, 4L, 5L,
8L, 9L, 15L, 26L, 36L, 34L, 2L, 4L, 5L, 8L, 9L, 15L, 26L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 5L, 8L, 9L, 36L, 34L, 2L, 4L, 5L, 8L,
9L, 36L, 34L, 2L, 4L, 5L),
no2_quintile = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L),
.Label = c("q1", "q2",
"q3", "q4", "q5"), class = "factor")),
class = "data.frame", row.names = c(NA,
-100L))
Any help is appreciated!