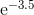Using the debugger, we can track the problem down to pairwise_comparisons, which is what throws the error:
pairwise_comparisons(data, day, measurement)
#> Error in data.frame(..., check.names = FALSE) :
#> arguments imply differing number of rows: 0, 1
And by debugging this function, we find that the error is thrown here:
df <- suppressWarnings(exec(.fn = .f, x = y_vec, g = x_vec, groups = x_vec,
blocks = g_vec, paired = paired, p.adjust.method = "none",
!!!.f.args)) %>% tidy_model_parameters(.) %>% rename(group2 = group1,
group1 = group2)
From examining these variables, we find that .f is PMCMRplus::gamesHowellTest, which although it does not throw an error, certainly doesn't like the input:
PMCMRplus::gamesHowellTest(data$measurement, g = data$day)
Pairwise comparisons using Games-Howell test
data: data$measurement and data$day
6 10 13
10 - - -
13 - - -
26 - - -
Essentially, it cannot carry out pairwise comparisons because there are not enough observations (only 2 in each group, which is insufficient to generate p values). Therefore when this result is passed to tidy_model_parameters(.), and the code tries to create a summary data frame including p values, the p value column is missing.
Ultimately it is therefore cbind that throws the error when the function attempts to bind a length-0 column to a length-1 column. We can replicate this exact error by trying to cbind a 1-row and 0-row data frame together:
cbind(data.frame(a = 1), data.frame(b = numeric()))
#> Error in data.frame(..., check.names = FALSE) :
#> arguments imply differing number of rows: 1, 0
So it is not your data frame, but the summary data frame of pairwise comparisons that has different row numbers. The obvious solution is to not attempt pairwise comparisons with such a small number of observations:
ggbetweenstats(data = df,
x = day,
y = measurement,
pairwise.comparisons = FALSE)
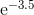
Note also that if we give it enough data points (even just by binding two copies of your data frame together), we get a pairwise comparison plot without any errors:
ggbetweenstats(data = rbind(df, df),
x = day,
y = measurement)

To be fair, this error is essentially uncaught by ggstatsplot, since there is no specific message to tell you what caused it or why, and it takes a bit of digging to discover the problem. Might be worth filing a bug report on the ggstatplot github page?