This is the dput output.
structure(list(data = structure(list(stations = c("s1", "s2",
"s3", "s4", "s5", "s1", "s2", "s3", "s4", "s5", "s1", "s2", "s3",
"s4", "s5", "s1", "s2", "s3", "s4", "s5"), Mean = c(792.0666667,
830.0666667, 542.4666667, 311.3333333, NA, 535.3222222, 576.9855556,
510.0756667, 433.9747778, 347.35, 726.9027778, 798.3153333,
706.2138889, 593.0828889, 474.9132222, 991.5945841, 1044.328044,
693.3808187, 403.8107808, NA), stdev = c(189.6764965, 123.9226103,
115.742876, 70.03437251, NA, 300.4539788, 306.3421887, 274.6515927,
237.7002122, 191.230147, 197.9449891, 248.458886, 203.0634826,
156.500875, 124.8746002, 265.4692578, 190.0975192, 185.9667516,
119.4178709, NA), Lower = c(602.3901701, 706.1440564, 426.7237907,
241.2989608, NA, 234.8682435, 270.6433669, 235.424074, 196.2745655,
156.119853, 528.9577886, 549.8564473, 503.1504063, 436.5820139,
350.038622, 726.1253263, 854.2305244, 507.4140671, 284.3929099, NA),
Upper = c(981.7431632, 953.9892769, 658.2095426, 381.3677058, NA,
835.776201, 883.3277442, 784.7272594, 671.67499, 538.580147,
924.8477669, 1046.774219, 909.2773715, 749.5837639, 599.7878224,
1257.063842, 1234.425563, 879.3475703, 523.2286518, NA), Group =
c("Observation", "Observation", "Observation", "Observation",
"Observation", "Historical", "Historical", "Historical", "Historical",
"Historical", "Future", "Future", "Future", "Future", "Future",
"Downscaled", "Downscaled", "Downscaled", "Downscaled", "Downscaled")),
class = "data.frame", row.names = c(NA, -20L)), layers =
list(<environment>, <environment>), scales = <environment>,
mapping = structure(list(x = ~Mean, Group = ~Group), class =
"uneval"), theme = list(), coordinates = <environment>, facet =
<environment>, plot_env = <environment>, labels = list(title =
"Probability density functions (PDF) for in comparison with
observations", y = "Probability density function", x = "March maximum
temperature", Group = "Group", colour = "factor(Group)", linetype =
"factor(Group)", fill = structure("fill", fallback = TRUE), weight =
structure("weight", fallback = TRUE))), class = c("gg",
"ggplot"))
I have produced density plot using ggplot2. The density plot is the mean. I want to shade each density line by lower and upper bounds just like confidence intervals. Please can anyone help how to achieve this?
The code I used to generate density in ggplot
ggplot(pdf1, aes(x=Mean, Group=Group)) +
geom_density(aes(colour=factor(Group), linetype=factor(Group)),
show.legend = FALSE) +
stat_density(aes(x=Mean, colour=factor(Group),
linetype=factor(Group)),
geom="line",position="identity", linewidth=1) +
xlab("March maximum temperature") +
ylab("Probability density function") +
ggtitle("Probability density functions (PDF) for in comparison with
observations") +
scale_linetype_manual(values=c('solid', 'solid','solid','dotted'),
labels=c("Observation", "Historical", "Future",
"Downscaled")) +
scale_colour_manual(values = c("black", "green", "blue", "red"),
labels=c("Observation", "Historical", "Future",
"Downscaled"))
This is the output from the above code 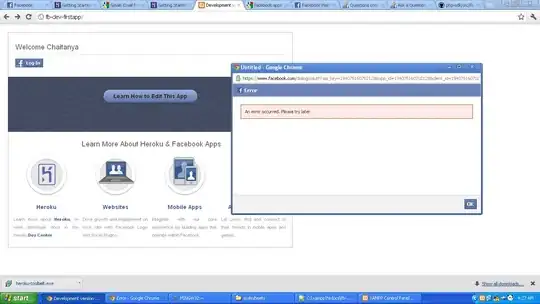
I searched in the net and could not find the solution.
Thanks a lot for your help!
This is a kind of output I am looking for (https://rpubs.com/Grady/875225). But as you can see, it has both x and y variables but I only have one variable (Mean in my case) in the x-axis while it is probability density in the y-axis in the above ggplot. So, I want to use the lower and upper values to envelope each line with shades with each respective color of my density plot