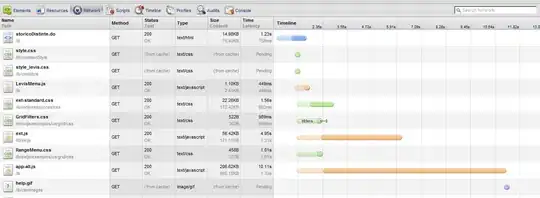
I applied different treatments to my samples and subsequently measured them at two different temperatures. Now I want to visualise the paired observations for each sample by adding lines between the two points beloning to one sample using ggplot2. However, when I try this, the lines do not connect the two points but appear as vertical lines in my plot. EDIT: Regarding the duplicate, I think the main difference with my data is I have more than two categories in the variable "fishid".
This is my data:
dput(s2n)
structure(list(fishid = structure(c(1L, 1L, 2L, 2L, 3L, 3L, 4L,
4L, 5L, 5L, 6L, 6L, 7L, 7L, 8L, 8L, 80L, 80L, 81L, 81L, 82L,
82L, 83L, 83L, 84L, 84L, 85L, 85L, 86L, 86L), .Label = c("1",
"2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13",
"14", "15", "16", "17", "18", "19", "20", "21", "22", "23", "24",
"25", "26", "27", "28", "29", "30", "31", "32", "33", "34", "35",
"36", "37", "38", "39", "40", "41", "42", "43", "44", "45", "47",
"48", "49", "50", "51", "52", "53", "54", "55", "56", "57", "58",
"59", "60", "61", "62", "63", "64", "65", "66", "67", "68", "69",
"70", "71", "72", "73", "74", "75", "76", "77", "78", "79", "80",
"81", "82", "83", "84", "85", "86", "87", "88", "89", "90", "91",
"92", "93", "94", "95", "96", "97", "98", "99", "100", "101",
"102", "103", "104", "105", "107", "108", "109", "110", "111",
"112", "113", "114"), class = "factor"), mtemp = c("23", "28",
"23", "28", "23", "28", "23", "28", "23", "28", "23", "28", "23",
"28", "23", "28", "23", "28", "23", "28", "23", "28", "23", "28",
"23", "28", "23", "28", "23", "28"), treat = c("2n 28 28 n",
"2n 28 28 n", "2n 28 28 n", "2n 28 28 n", "2n 28 28 n", "2n 28 28 n",
"2n 28 28 n", "2n 28 28 n", "2n 28 28 n", "2n 28 28 n", "2n 28 28 n",
"2n 28 28 n", "2n 28 28 n", "2n 28 28 n", "2n 28 28 n", "2n 28 28 n",
"2n 23 23 n", "2n 23 23 n", "2n 23 23 n", "2n 23 23 n", "2n 23 23 n",
"2n 23 23 n", "2n 23 23 n", "2n 23 23 n", "2n 23 23 n", "2n 23 23 n",
"2n 23 23 n", "2n 23 23 n", "2n 23 23 n", "2n 23 23 n"), N = c(4,
3, 4, 4, 3, 3, 4, 4, 4, 3, 4, 4, 4, 4, 4, 4, 3, 1, 4, 4, 3, 4,
3, 4, 3, 4, 1, 2, 1, 4), mo2_ms = c(0.0305521553416736, 0.145682463746226,
0.0396710467863138, 0.0954773661923973, 0.0423458775047231, 0.074814122089128,
0.0340000306491368, 0.164506483241493, 0.0199183915444751, 0.0141578076646164,
0.0352100227426512, 0.140223575206394, 0.0867953316734817, 0.181380336846186,
0.0814477532477152, 0.245441716187935, 0.0753519288551872, 0.192765607492084,
0.112553420128519, 0.154874683462045, 0.096957746918988, 0.174162792054346,
0.0572348282730377, 0.198498172699998, 0.0969845912494747, 0.0883266097299638,
0.00529159243126301, 0.048338396851318, 0.0154151384427716, 0.165540613767348
), sd = c(0.0225825414793994, 0.040281910779122, 0.0353185196092258,
0.0524668087896432, 0.0229625978946772, 0.00715984733525644,
0.0213724448650181, 0.0416621727661058, 0.00851354415170029,
0.00409799422855184, 0.0147318782789288, 0.0800135890495444,
0.0207362723426385, 0.0171873141998392, 0.0526956334274908, 0.0569951399710065,
0.0352363626542992, NA, 0.0752014409258304, 0.018373804187377,
0.0294532805712796, 0.0188553911271472, 0.0414618539087027, 0.0294471142185835,
0.0120861972052966, 0.0268913613699161, NA, 0.0614419583639327,
NA, 0.0919378871844233), se = c(0.0112912707396997, 0.0232567720317986,
0.0176592598046129, 0.0262334043948216, 0.013257462075785, 0.00413373978636693,
0.0106862224325091, 0.0208310863830529, 0.00425677207585014,
0.00236597807099194, 0.00736593913946441, 0.0400067945247722,
0.0103681361713193, 0.00859365709991959, 0.0263478167137454,
0.0284975699855032, 0.0203437234637229, NA, 0.0376007204629152,
0.00918690209368851, 0.0170048594663459, 0.00942769556357362,
0.0239380125152904, 0.0147235571092918, 0.00697796920995688,
0.0134456806849581, NA, 0.0434460254085183, NA, 0.0459689435922116
), ci = c(0.0359338628475153, 0.100065813667779, 0.0561996461192354,
0.0834864008913134, 0.0570422553899265, 0.0177860467758856, 0.0340083290977783,
0.0662938138947925, 0.0135469485652242, 0.0101799820056808, 0.0234417057993332,
0.127319475421606, 0.0329960366510934, 0.0273488522865142, 0.0838505119529514,
0.0906919863099286, 0.0875319772944557, NaN, 0.11966227391321,
0.0292368226250621, 0.073166005001879, 0.0300031349136345, 0.102996954893691,
0.0468569299230992, 0.0300237782693287, 0.0427901568178693, NaN,
0.552034093813681, NaN, 0.146293694692829)), row.names = c(1L,
2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 14L, 15L,
16L, 150L, 151L, 152L, 153L, 154L, 155L, 156L, 157L, 158L, 159L,
160L, 161L, 162L, 163L), class = "data.frame")
This is the code for the plot I created without the connecting lines:
p1 <- ggplot(data = s2n, aes(x = treat, y = mo2_ms, fill = mtemp)) +
geom_boxplot() +
geom_point(position=position_jitterdodge(jitter.width = 0, jitter.height = 0)) # I am using the position_jitterdodge to actually get the points to align to the middel of the boxplots
p1
This results in the following plot:
I want to create a plot like this, but then with the two treatments within the same plot:
I tried using this code to add the connecting lines with the two treatments within the same plot:
p3 <- ggplot(data = s2n, aes(x = treat, y = mo2_ms, fill = mtemp)) +
geom_boxplot() +
geom_point(position=position_jitterdodge(jitter.width = 0, jitter.height = 0)) +
geom_line(aes(group=fishid))
p3
This is the resulting plot:
When I try using the same position_jitterdodge in the geom_line argument I get this:
p4 <- ggplot(data = s2n, aes(x = treat, y = mo2_ms, fill = mtemp)) +
geom_boxplot() +
geom_point(position=position_jitterdodge(jitter.width = 0, jitter.height = 0)) +
geom_line(position=position_jitterdodge(jitter.width = 0, jitter.height = 0), aes(group=fishid))
p4
Resulting in this plot:
Does anybody have some suggestions on how to get the lines to connect the points in plot4?