I'm not sure what you are trying to do. You haven't provided a reproducible dataset and it makes it pretty difficult for others to chip in and help without it. It also appears you are doing something somewhat odd, you're trying to change the predictor values and the estimated y values at the same time. The y-axis will necessarily change by virtue of changing the predictor values. What specifically is the purpose of this? To me this isn't super helpful, but perhaps you can elaborate in a comment here.
Instead of trying what you did, I have simulated some data to try to resemble yours, then fit a lme4 regression. Thereafter I plot an interaction with a different function, the plot_model function from the sjPlot package. First, the requisite packages are loaded below, one for the mixed models regression and one for the plotting function.
#### Load Libraries ####
library(lmerTest)
library(sjPlot)
Then I tried simulating your data to at least resemble what you described and created a data frame called df below.
#### Simulate Data ####
set.seed(123)
tree <- sample(seq(0,1.5,by=.01),
100)
ph <- sample(seq(3,9,by=.01),
100)
y <- rnorm(n=1000)
re <- factor(round(rbinom(n=1000,size=5,prob = .5)),
labels = c("site1","site2","site3","site4","site5","site6"))
df <- data.frame(tree,ph,y,re)
df
Then I fit the model to a linear mixed model with this code.
#### Fit Model ####
fit <- lmer(y ~ tree*ph + (1|re),
df)
summary(fit)
To plot the interactions for this, one can use the plot_model function mentioned previously. We can specify method = "int" to plot interactions, then specify mdrt.values = "meansd" to get it to plot the interactions using slices with the mean and one standard deviation above and below the mean. Of course you also select the terms with the interaction with the terms argument. Because it is a ggplot object, you can also add some extra customization of the labels, so I added an x/y axis name and title.
#### Plot ####
plot_model(fit,
type = "int",
mdrt.values = "meansd",
terms = c("tree","ph"))+
labs(x="Tree Size",
y="Growth Rate",
title = "Interaction Between pH Levels and Tree Size on Growth Rate")
Which should look like this:
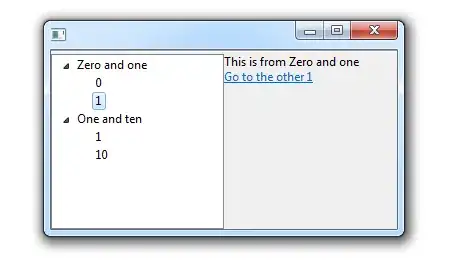
While this doesn't give you the exact values you want, it at least gives you meaningful ones. You now have values that resemble what z-score changes affect the direction of the interaction.