I am trying to connect the centroids of buildings (and other areas) to a NetworkX.MultiGraph representing the road network. These centroids are all 0-degree nodes.
I have read posts on finding the nearest neighbors in the same graph but failed to find anything for 'off-grid' nodes. Graph.neighbors() and ego_graph() all returned empty iterables for the centroid nodes.
I wonder if there is a function for this case. Do I have to iterate over all the nodes on the roads to compute their distances to the centroid? Or there is a better idea?
The red dots are the nodes to connect.
Minimal example data
some_edges = [((12965572.317313856, 4069201.5797910197),
(12965582.853703659, 4069201.5541923903)),
((12965582.853703659, 4069201.5541923903),
(12965593.39009346, 4069201.528593761)),
((12965593.39009346, 4069201.528593761),
(12965637.645157026, 4069201.426199232)),
((12965593.39009346, 4069201.528593761),
(12965594.291781338, 4069120.115297204)),
((12965593.39009346, 4069201.528593761),
(12965593.790843628, 4069233.4285842725))]
a_centroid = [((12965783.891266523, 4069098.4402361237), {'id': 839812059, 'node_type': 'area_centroid', 'amenity': 'library', 'building': 'yes', 'name': '逸夫图书馆', 'name:zh': '逸夫图书馆', 'name:zh-Hans': '逸夫图书馆', 'name:zh-Hant': '逸夫圖書館', 'operator': '北京工业大学'})]
G = nx.MultiGraph(some_edges)
G.add_nodes_from(a_centroid)
color_map = ['red' if 'name' in attr.keys() else 'blue' for n, attr in G.nodes.data()]
nx.draw(G, {n: (n[0], n[1]) for n in G.nodes()}, node_color=color_map, ax=plt.gca())
should produce a minimal example with a graph like this:
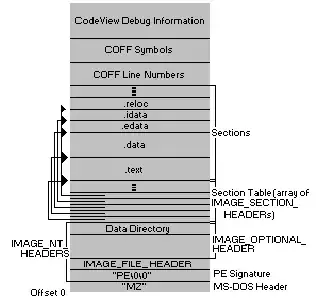
where the blue nodes are parts of the graph while the red node is a centroid to connect (to its nearest node in the graph).