Let's say I have dataset within the following pandas dataframe format with a non-standard timestamp column without datetime format as follows:
+--------+-----+
|TS_24hrs|count|
+--------+-----+
|0 |157 |
|1 |334 |
|2 |176 |
|3 |86 |
|4 |89 |
... ...
|270 |192 |
|271 |196 |
|270 |251 |
|273 |138 |
+--------+-----+
274 rows × 2 columns
I have already applied some regression algorithms after splitting data without using cross-validation (CV) into training-set and test-set and got results like the following:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
#Load the time-series data as dataframe
df = pd.read_csv('/content/U2996_24hrs_.csv', sep=",")
print(df.shape)
# Split the data into training and testing sets
from sklearn.model_selection import train_test_split
train, test = train_test_split(df, test_size=0.27, shuffle=False)
print(train.shape) #(200, 2)
print(test.shape) #(74, 2)
#visulize splitted data
train['count'].plot(label='Training-set')
test['count'].plot(label='Test-set')
plt.legend()
plt.show()
#Train and fit the model
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor().fit(train, train['count']) #X, y
rf.score(train, train['count']) #0.9998644192184375
# Use the forest's model to predict on the test-set
predictions = rf.predict(test)
#convert prediction result into dataframe for plot issue in ease
df_pre = pd.DataFrame({'TS_24hrs':test['TS_24hrs'], 'count_prediction':predictions})
# Calculate the mean absolute errors
from sklearn.metrics import mean_absolute_error
rf_mae = mean_absolute_error(test['count'], df_pre['count_prediction'])
print(train.shape) #(200, 2)
print(test.shape) #(74, 2)
print(df_pre.shape) #(74, 2)
#visulize forecast or prediction of used regressor model
train['count'].plot(label='Training-set')
test['count'].plot(label='Test-set')
df_pre['count_prediction'].plot(label=f'RF_forecast MAE={rf_mae:.2f}')
plt.legend()
plt.show()
According this answer I noticed:
if your data is already sorted based on time then simply use
shuffle=Falseintrain, test = train_test_split(newdf, test_size=0.3, shuffle=False)
So far, I have used this classic split data method, but I want to experiment with Time-series-based split methods that are summarized here:
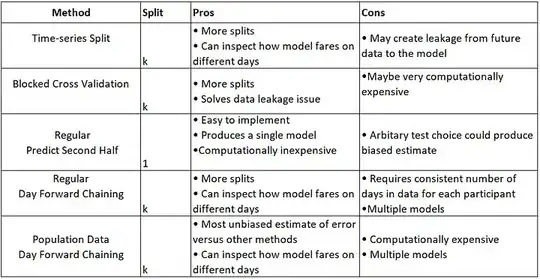
Additionally, based on my investigation (please see the references at the end of the post), it is recommended to use the cross-validation method (K-Fold) before applying regression models. explanation: Cross Validation in Time Series
Problem: How can split time-series data with using CV methods for comparable results? (plot the quality of data split for ensure\evaluate the quality of data splitting)
- TSS CV method:
TimeSeriesSplit() - BTSS CV method:
BlockingTimeSeriesSplit()
So far, the closest solution that crossed my mind is to separate the last 74 observations as hold-on test-set a side and do CV on just the first 200 observations. I'm still struggling with playing with these arguments max_train_size=199, test_size=73 to reach desired results, but it's very tricky and I couldn't figure it out. in fact, I applied time-series-based data split using TSS CV methods before training RF regressor to train-set (first 200 days\observations) and fit model over test-set (last 74 days\observations).
I've tried recommended TimeSeriesSplit() as the following unsuccessfully:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
#Load the time-series data as dataframe
df = pd.read_csv('/content/U2996_24hrs_.csv', sep=",")
print(df.shape)
#Try to split data with CV (K-Fold) by using TimeSeriesSplit() method
from sklearn.model_selection import TimeSeriesSplit
tscv = TimeSeriesSplit(
n_splits=len(df['TS_24hrs'].unique()) - 1,
gap=0, # since data alraedy groupedby for 24hours to retrieve daily count there is no need to to have gap
#max_train_size=199, #here: https://stackoverflow.com/a/43326651/10452700 they recommended to set this argument I'm unsure if it is the case for my problem
#test_size=73,
)
for train_idx, test_idx in tscv.split(df['TS_24hrs']):
print('TRAIN: ', df.loc[df.index.isin(train_idx), 'TS_24hrs'].unique(),
'val-TEST: ', df.loc[df.index.isin(test_idx), 'TS_24hrs'].unique())
The following figures for understanding and better alignment of split data could be part of the expected output if one could plot for each method:
expected output:
References:
Using k-fold cross-validation for time-series model selection
Cross Validation for Time Series Classification (Not Forecasting!)
Edit1:
I found 3 related posts:
- post1
- post2
I decided to apply
TimeSeriesSplit()in short TTS cv output within for loop to train\fit regression model over training-set with assist of CV-set thenpredict()over Hold-on test-set. The current output of my implementation shows slightly improvement in forecasting with or without, which could be due to problems in my implementation.
#Load the time-series data as dataframe
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('/content/U2996_24hrs_.csv', sep=",")
#print(df.shape) #(274, 2)
#####----------------------------without CV
# Split the data into training and testing sets
from sklearn.model_selection import train_test_split
train, test = train_test_split(df, test_size=0.27, shuffle=False)
print(train.shape) #(200, 2)
print(test.shape) #(74, 2)
#visulize splitted data
#train['count'].plot(label='Training-set')
#test['count'].plot(label='Test-set')
#plt.legend()
#plt.show()
#Train and fit the model
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor().fit(train, train['count']) #X, y
rf.score(train, train['count']) #0.9998644192184375
# Use the forest's model to predict on the test-set
predictions = rf.predict(test)
#convert prediction result into dataframe for plot issue in ease
df_pre = pd.DataFrame({'TS_24hrs':test['TS_24hrs'], 'count_prediction':predictions})
# Calculate the mean absolute errors
from sklearn.metrics import mean_absolute_error
rf_mae = mean_absolute_error(test['count'], df_pre['count_prediction'])
#####----------------------------with CV
df1 = df[:200] #take just first 1st 200 records
#print(df1.shape) #(200, 2)
#print(len(df1)) #200
from sklearn.model_selection import TimeSeriesSplit
tscv = TimeSeriesSplit(
n_splits=len(df1['TS_24hrs'].unique()) - 1,
#n_splits=3,
gap=0, # since data alraedy groupedby for 24hours to retrieve daily count there is no need to to have gap
#max_train_size=199,
#test_size=73,
)
#print(type(tscv)) #<class 'sklearn.model_selection._split.TimeSeriesSplit'>
#mae = []
cv = []
TS_24hrs_tss = []
predictions_tss = []
for train_index, test_index in tscv.split(df1):
cv_train, cv_test = df1.iloc[train_index], df1.iloc[test_index]
#cv.append(cv_test.index)
#print(cv_train.shape) #(199, 2)
#print(cv_test.shape) #(1, 2)
TS_24hrs_tss.append(cv_test.values[:,0])
#Train and fit the model
from sklearn.ensemble import RandomForestRegressor
rf_tss = RandomForestRegressor().fit(cv_train, cv_train['count']) #X, y
# Use the forest's model to predict on the cv_test
predictions_tss.append(rf_tss.predict(cv_test))
#print(predictions_tss)
# Calculate the mean absolute errors
#from sklearn.metrics import mean_absolute_error
#rf_tss_mae = mae.append(mean_absolute_error(cv_test, predictions_tss))
#print(rf_tss_mae)
#print(len(TS_24hrs_tss)) #199
#print(type(TS_24hrs_tss)) #<class 'list'>
#print(len(predictions_tss)) #199
#convert prediction result into dataframe for plot issue in ease
import pandas as pd
df_pre_tss1 = pd.DataFrame(TS_24hrs_tss)
df_pre_tss1.columns =['TS_24hrs_tss']
#df_pre_tss1
df_pre_tss2 = pd.DataFrame(predictions_tss)
df_pre_tss2.columns =['count_predictioncv_tss']
#df_pre_tss2
df_pre_tss= pd.concat([df_pre_tss1,df_pre_tss2], axis=1)
df_pre_tss
# Use the forest's model to predict on the hold-on test-set
predictions_tsst = rf_tss.predict(test)
#print(len(predictions_tsst)) #74
#convert prediction result of he hold-on test-set into dataframe for plot issue in ease
df_pre_test = pd.DataFrame({'TS_24hrs_tss':test['TS_24hrs'], 'count_predictioncv_tss':predictions_tsst})
# Fix the missing record (1st record)
df_col_merged = df_pre_tss.merge(df_pre_test, how="outer")
#print(df_col_merged.shape) #(273, 2) 1st record is missing
ddf = df_col_merged.rename(columns={'TS_24hrs_tss': 'TS_24hrs', 'count_predictioncv_tss': 'count'})
df_first= df.head(1)
df_merged_pred = df_first.merge(ddf, how="outer") #insert first record from original df to merged ones
#print(df_merged_pred.shape) #(274, 2)
print(train.shape) #(200, 2)
print(test.shape) #(74, 2)
print(df_pre_test.shape) #(74, 2)
# Calculate the mean absolute errors
from sklearn.metrics import mean_absolute_error
rf_mae_tss = mean_absolute_error(test['count'], df_pre_test['count_predictioncv_tss'])
#visulize forecast or prediction of used regressor model
train['count'].plot(label='Training-set', alpha=0.5)
test['count'].plot(label='Test-set', alpha=0.5)
#cv['count'].plot(label='cv TSS', alpha=0.5)
df_pre['count_prediction'].plot(label=f'RF_forecast MAE={rf_mae:.2f}', alpha=0.5)
df_pre_test['count_predictioncv_tss'].plot(label=f'RF_forecast_tss MAE={rf_mae_tss:.2f}', alpha=0.5 , linestyle='--')
plt.legend()
plt.title('Plot forecast results with & without cross-validation (K-Fold)')
plt.show()
- post3 sklearn
(I couldn't implement it, one can try this) using
make_pipeline()and usedef evaluate(model, X, y, cv):function but still confusing if I want to collect the results in the form of dataframe for visualizing case and what is the best practice to pass cv result to regressor and compare the results.
Edit2: In the spirit of DRY, I tried to build an end-to-end pipeline without/with CV methods, load a dataset, perform feature scaling and supply the data into a regression model:
#Load the time-series data as dataframe
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('/content/U2996_24hrs_.csv', sep=",")
#print(df.shape) #(274, 2)
#####--------------Create pipeline without CV------------
# Split the data into training and testing sets for just visualization sense
from sklearn.model_selection import train_test_split
train, test = train_test_split(df, test_size=0.27, shuffle=False)
print(train.shape) #(200, 2)
print(test.shape) #(74, 2)
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.ensemble import RandomForestRegressor
from sklearn.pipeline import Pipeline
# Split the data into training and testing sets without CV
X = df['TS_24hrs'].values
y = df['count'].values
print(X_train.shape) #(200, 1)
print(y_train.shape) #(200,)
print(X_test.shape) #(74, 1)
print(y_test.shape) #(74,)
# Here is the trick
X = X.reshape(-1,1)
X_train, X_test, y_train, y_test = train_test_split(X, y , test_size=0.27, shuffle=False, random_state=0)
print(X_train.shape) #(200, 1)
print(y_train.shape) #(1, 200)
print(X_test.shape) #(74, 1)
print(y_test.shape) #(1, 74)
#build an end-to-end pipeline, and supply the data into a regression model. It avoids leaking the test set into the train set
rf_pipeline = Pipeline([('scaler', MinMaxScaler()),('RF', RandomForestRegressor())])
rf_pipeline.fit(X_train, y_train)
#Displaying a Pipeline with a Preprocessing Step and Regression
from sklearn import set_config
set_config(display="diagram")
rf_pipeline # click on the diagram below to see the details of each step
r2 = rf_pipeline.score(X_test, y_test)
print(f"RFR: {r2}") # -0.3034887940244342
# Use the Randomforest's model to predict on the test-set
y_predictions = rf_pipeline.predict(X_test.reshape(-1,1))
#convert prediction result into dataframe for plot issue in ease
df_pre = pd.DataFrame({'TS_24hrs':test['TS_24hrs'], 'count_prediction':y_predictions})
# Calculate the mean absolute errors
from sklearn.metrics import mean_absolute_error
rf_mae = mean_absolute_error(y_test, df_pre['count_prediction'])
print(train.shape) #(200, 2)
print(test.shape) #(74, 2)
print(df_pre.shape) #(74, 2)
#visulize forecast or prediction of used regressor model
train['count'].plot(label='Training-set')
test['count'].plot(label='Test-set')
df_pre['count_prediction'].plot(label=f'RF_forecast MAE={rf_mae:.2f}')
plt.legend()
plt.title('Plot results without cross-validation (K-Fold) using pipeline')
plt.show()
#####--------------Create pipeline with TSS CV------------
#####--------------Create pipeline with BTSS CV------------
The results got worse using the pipeline, based on MAE score comparing implementation when separating the steps outside of the pipeline!
 Note: I calculate the mean of splits (K-folds):
Note: I calculate the mean of splits (K-folds):