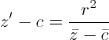This is a common problem with PDF and tabular text
Left is the PDF rendered layout - Middle is raw extraction order - Right is what we expect (from a clipboard output that uses screen layout)

What we need is closest to this (still not perfect due to alignment and missing ≥ symbol)
Organ Dose (mGy/MBq) E.2.2. Imaging devices:
Adults Bladder 0.05 0.0077 - Multiple detector (triple or dual head) or other
99mTc-ECDa Kidney 0.034 0.0093 dedicated small to medium field of view
99mTc-HMPAOb SPECT cameras for brain imaging should be
Bladder 0.11 0.022 used for acquisition since these devices gen-
Children (5 years) Thyroid 0.14 0.027 erally produce results superior to those
99mTc-ECDa obtained with single-head cameras.
99mTc-HMPAOb
- Single-detector units may only be used if
a ICRP 106, page 107 the scan time is prolonged appropriately (so
b ICRP 80, page 100 as to reach at least 5 million total detected
slightly better
Organ Dose (mGy/MBq) dedicated small to medium field of view
Adults SPECT cameras for brain imaging should be
99mTc-ECDa Bladder 0.05 0.0077 used for acquisition since these devices gen-
99mTc-HMPAOb Kidney 0.034 0.0093 erally produce results superior to those
Children (5 years) obtained with single-head cameras.
99mTc-ECDa Bladder 0.11 0.022 - Single-detector units may only be used if
99mTc-HMPAOb Thyroid 0.14 0.027 the scan time is prolonged appropriately (so
as to reach at least 5 million total detected
aICRP 106, page 107 events) and meticulous care is taken to
bICRP 80, page 100 produce high-quality images.
so the best command is pdftotext -layout
and we now have a perfect working input to start parsing in python as text
For Windows Command Line Instructions (CLI) to convert to CSV its a second command line or so, for a simple case it could be (different source) where we desire to abbreviate column headers and at the same time, exclude the x symbol/column

@pdftotext -nopgbrk -f 1 -l 1 -layout -x 290 -y 530 -W 300 -H 300 cut-sample.pdf out.txt
@echo Pc,W,H,Q,C>out.csv&for /f "usebackq tokens=1,2,4,5,6 delims= " %%f in ("out.txt") do @echo %%f,%%g,%%h,%%i,%%j >>out.csv
@echo/&type out.csv
However for this type of layout we need a different approach depending on how much is wanted,
@echo off
echo "Range (µm)",2.00,≥ 10.00,≥ 25.00,≥ 50.00,≥ 100.00 >out.csv
echo " ",^< 10.00,^< 1751.00,^< 1751.00,^< 1751.00,^< 1751.00 >>out.csv
for /f "usebackq tokens=1,2,3,4,5,6,7 delims= " %%A in ("particles.txt") do (
if "%%A"=="Count" echo %%A,%%B,%%C,%%D,%%E,%%F >>out.csv
if "%%A"=="-" echo " ",%%A,%%B,%%C,%%D,%%E >>out.csv
if "%%A"=="Concentration" echo %%A %%B,%%C,%%D,%%E,%%F,%%G >>out.csv
)
echo/&"C:\Apps\Microsoft Office\Office\Excel" out.csv
However note that csv as 7/8bit text and UTF-8 symbols do not work well without a pure 8bit format. The CSV is perfect 8 bits WinAnsi and Notepad can save as UTF-8, but older Excel expects Ascii (lower 7 bits of Ansi)