If you call leastsq like this:
import scipy.optimize
p,cov,infodict,mesg,ier = optimize.leastsq(
residuals,a_guess,args=(x,y),full_output=True)
where
def residuals(a,x,y):
return y-f(x,a)
then, using the definition of R^2 given here,
ss_err=(infodict['fvec']**2).sum()
ss_tot=((y-y.mean())**2).sum()
rsquared=1-(ss_err/ss_tot)
What is infodict['fvec'] you ask? It's the array of residuals:
In [48]: optimize.leastsq?
...
infodict -- a dictionary of optional outputs with the keys:
'fvec' : the function evaluated at the output
For example:
import scipy.optimize as optimize
import numpy as np
import collections
import matplotlib.pyplot as plt
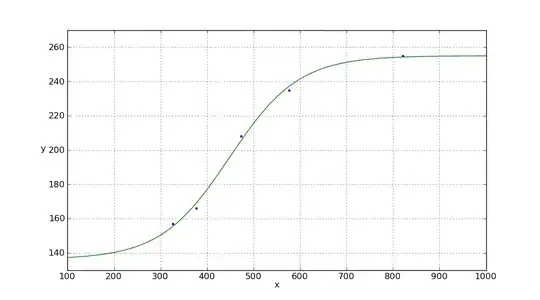
x = np.array([821,576,473,377,326])
y = np.array([255,235,208,166,157])
def sigmoid(p,x):
x0,y0,c,k=p
y = c / (1 + np.exp(-k*(x-x0))) + y0
return y
def residuals(p,x,y):
return y - sigmoid(p,x)
Param=collections.namedtuple('Param','x0 y0 c k')
p_guess=Param(x0=600,y0=200,c=100,k=0.01)
p,cov,infodict,mesg,ier = optimize.leastsq(
residuals,p_guess,args=(x,y),full_output=True)
p=Param(*p)
xp = np.linspace(100, 1600, 1500)
print('''\
x0 = {p.x0}
y0 = {p.y0}
c = {p.c}
k = {p.k}
'''.format(p=p))
pxp=sigmoid(p,xp)
# You could compute the residuals this way:
resid=residuals(p,x,y)
print(resid)
# [ 0.76205302 -2.010142 2.60265297 -3.02849144 1.6739274 ]
# But you don't have to compute `resid` -- `infodict['fvec']` already
# contains the info.
print(infodict['fvec'])
# [ 0.76205302 -2.010142 2.60265297 -3.02849144 1.6739274 ]
ss_err=(infodict['fvec']**2).sum()
ss_tot=((y-y.mean())**2).sum()
rsquared=1-(ss_err/ss_tot)
print(rsquared)
# 0.996768131959
plt.plot(x, y, '.', xp, pxp, '-')
plt.xlim(100,1000)
plt.ylim(130,270)
plt.xlabel('x')
plt.ylabel('y',rotation='horizontal')
plt.grid(True)
plt.show()