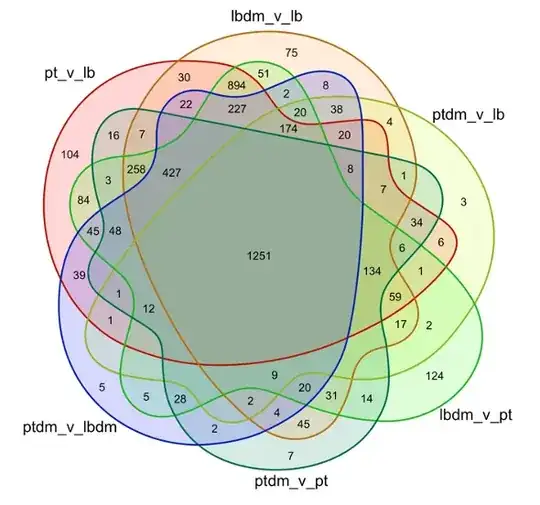
I have 6 lists. Each contains genes of interest from a given condition. These six lists were then used to make a venn diagram in r using the venn package.
The venn diagram is precisely what I want, but now I would like to produce a list of genes from each section of the venn diagram. how do I do this? i.e. how do I figure out what genes from my lists comprise the 1251 genes that are present in each list/condition?
library(venn)
venn_list_no <- list(pt_v_lb = rownames(pt_v_lb_no), lbdm_v_lb = rownames(lbdm_v_lb_no), ptdm_v_lb = rownames(ptdm_v_lb_no), lbdm_v_pt = rownames(lbdm_v_pt_no), ptdm_v_pt = rownames(ptdm_v_pt_no), ptdm_v_lbdm = rownames(ptdm_v_lbdm_no))
venn(venn_list_no, zcolor = "style", ggplot = T, opacity = 0.2, box = F)+
theme()