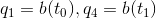Interesting question; one potential option would be to take the first non-NA value for each group, i.e.
Example data:
library(tidyverse)
data(ChickWeight)
df <- ChickWeight %>%
slice_head(n = 36) %>%
mutate(Diet = ifelse(Chick == 1, NA, Diet),
Diet = ifelse(Chick == 2 & Time < 16, NA, Diet))
df
#> weight Time Chick Diet
#> 1 42 0 1 NA
#> 2 51 2 1 NA
#> 3 59 4 1 NA
#> 4 64 6 1 NA
#> 5 76 8 1 NA
#> 6 93 10 1 NA
#> 7 106 12 1 NA
#> 8 125 14 1 NA
#> 9 149 16 1 NA
#> 10 171 18 1 NA
#> 11 199 20 1 NA
#> 12 205 21 1 NA
#> 13 40 0 2 NA
#> 14 49 2 2 NA
#> 15 58 4 2 NA
#> 16 72 6 2 NA
#> 17 84 8 2 NA
#> 18 103 10 2 NA
#> 19 122 12 2 NA
#> 20 138 14 2 NA
#> 21 162 16 2 1
#> 22 187 18 2 1
#> 23 209 20 2 1
#> 24 215 21 2 1
#> 25 43 0 3 1
#> 26 39 2 3 1
#> 27 55 4 3 1
#> 28 67 6 3 1
#> 29 84 8 3 1
#> 30 99 10 3 1
#> 31 115 12 3 1
#> 32 138 14 3 1
#> 33 163 16 3 1
#> 34 187 18 3 1
#> 35 198 20 3 1
#> 36 202 21 3 1
Answer:
# Chick 2 has both value and NA in reframed df
df %>%
reframe(Diet = unique(Diet), .by = Chick)
#> Chick Diet
#> 1 1 NA
#> 2 2 NA
#> 3 2 1
#> 4 3 1
# Potential solution
df %>%
reframe(Diet = first(unique(Diet), na_rm = TRUE), .by = Chick)
#> Chick Diet
#> 1 1 NA
#> 2 2 1
#> 3 3 1
Created on 2023-04-24 with reprex v2.0.2
Does this approach solve your problem?