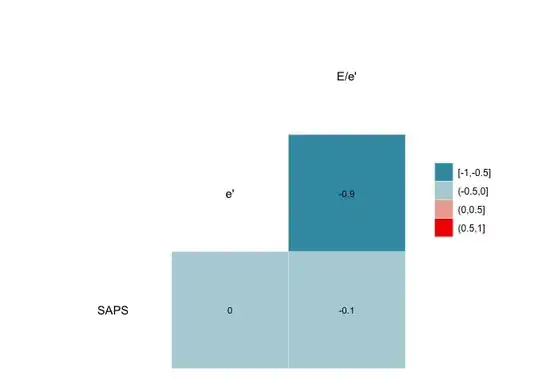
I ran into an issue, running the ggcorr package. My dataframe is quite simple, but column labels contain special characters:
| SAPS | e' | E/e' |
|---|---|---|
| 11 | 11,9 | 5,0 |
| 14 | 6,2 | 11,1 |
| 14 | 7,4 | 8,2 |
| 15 | 7,5 | 6,8 |
| 14 | 11,1 | 7,8 |
| 13 | 6,6 | 10,5 |
| 14 | 10,2 | 6,0 |
| 13 | 7,1 | 9,1 |
| 12 | 10,0 | 6,1 |
| 15 | 10,8 | 4,9 |
When I run the code as is, the output does not label correctly the variables. I wish to transpose the column names as is. In this example, the figure says e. instead of e' and E.e. instead of E/e'
How should I do it? Thank you in advance,
ggcorr(ex_db, nbreaks = 4,
label = TRUE,
label_size = 3,
method = c("pairwise", "spearman"))
I tried
ggcorr(ex_db, aes(x, y=c(SAPS, e', E/e')), method = c("pairwise", "spearman")) + geom_point()
without success