I currently use R to connect to a sql database and pull information. One particular code I use in R looks like this:
Joe<-dbGetQuery(jdbcConnection,"SELECT priv_check.protocol_id
FROM LHS_ONCORE_PROD.sv_user_pcl_permission priv_check
WHERE priv_check.function_name = 'CRPT-Subject Visits'
AND priv_check.contact_id = '6017'" )
Its a query that returns a dataframe of what protocols Joe (contact id 6017) is allowed by our clinical trial system to see. This dataframe is currently 1 column and ~500 rows. Kinda looks like this:
What I'd love is to create a very large dataframe that returns the protocols EVERY user can see.
Lets say I have a separate dataframe with the contact id's of our users, like this:
df<-structure(list(Contact = c("John", "Ali", "Joe", "Steve"), ContactID = c(1234,
4321, 6017, 5522)), row.names = c(NA, -4L), spec = structure(list(
cols = list(Contact = structure(list(), class = c("collector_character",
"collector")), ContactID = structure(list(), class = c("collector_double",
"collector"))), default = structure(list(), class = c("collector_guess",
"collector")), delim = ","), class = "col_spec"), class = c("spec_tbl_df",
"tbl_df", "tbl", "data.frame"))
and instead of just returning the dataframes joe (6017) can view, it also checks john (1234) and ali (4321) and steve (5522).
I realize this query will probably take a very long time to run, and I also think I'll have to figure out how to organize it when I get the data (in terms of long vs wide, etc...) but to begin with, I dont even know how to get it to automatically loop like that. Any ideas?
If possible, I'd love the output to line studies up as much as possible and look like this: 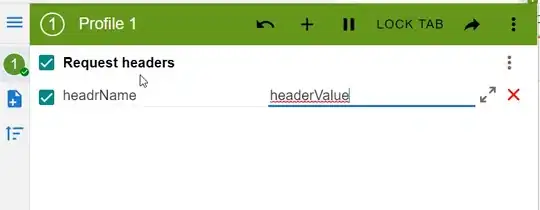
but thats not SUPER important