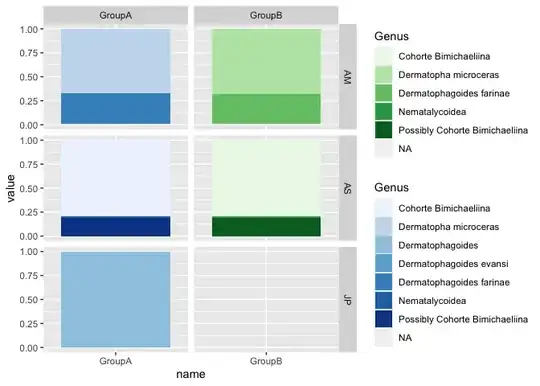
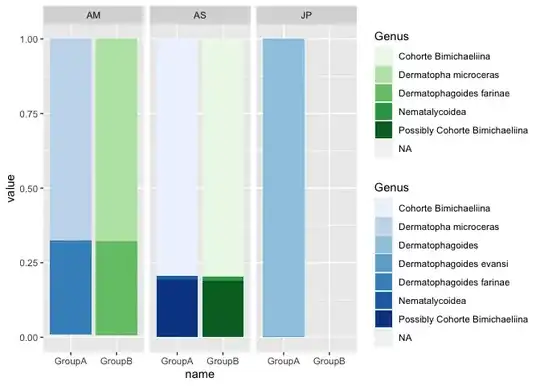
I have some problems with making a stacked bar chart in ggplot2.Here is my data look like:
ID Genus name value
1 JP Dermatophagoides GroupA 0.997
2 JP Dermatophagoides GroupB NA
3 JP Dermatophagoides evansi GroupA 0.00259
4 JP Dermatophagoides evansi GroupB NA
5 AM Dermatophagoides farinae GroupA 0.302
6 AM Dermatophagoides farinae GroupB 0.303
7 AM Dermatopha microceras GroupA 0.650
8 AM Dermatopha microceras GroupB 0.653
9 AM N/A GroupA 0.00915
10 AM N/A GroupB 0.00642
11 AS Cohorte Bimichaeliina GroupA 0.795
12 AS Cohorte Bimichaeliina GroupB 0.796
13 AS Nematalycoidea GroupA 0.0120
14 AS Nematalycoidea GroupB 0.0123
15 AS Possibly Cohorte Bimichaeliina GroupA 0.193
16 AS Possibly Cohorte Bimichaeliina GroupB 0.191
I am trying to visualize Genus,name and value based on every ID.I also tried the code at this page Showing data values on stacked bar chart in ggplot2, like I have tried :
df1 <- df %>% select(ID == "JP")
ggplot(dt1, aes(name, value, fill = Genus)) +
geom_col()
Desired format of plots like
But I only can handle one ID each time.I have multiple IDs,also want colored by name, for example GroupA using blue(dark to light) and GroupB using another Green(dark to light).Any help in this regard will be highly appreciated. Thanks.