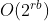Using R, I tried to create these two random shapefiles that represent two sets of "zones" within some city:
set.seed(123)
# Load the ggplot2 and sf packages
library(ggplot2)
library(sf)
# Set the longitude and latitude for New York
lon_1 <- -74.0060
lat_1 <- 40.7128
# Set the longitude and latitude for a second location
lon_2 <- -74.037
lat_2 <- 40.733
# Set the size of the square
size <- 0.1
# Create a square centered on New York
square_1 <- st_sfc(st_polygon(list(rbind(
c(lon_1 - size/2, lat_1 - size/2),
c(lon_1 + size/2, lat_1 - size/2),
c(lon_1 + size/2, lat_1 + size/2),
c(lon_1 - size/2, lat_1 + size/2),
c(lon_1 - size/2, lat_1 - size/2)
))))
# Create a square centered on New York
square_2 <- st_sfc(st_polygon(list(rbind(
c(lon_2 - size/2, lat_2 - size/2),
c(lon_2 + size/2, lat_2 - size/2),
c(lon_2 + size/2, lat_2 + size/2),
c(lon_2 - size/2, lat_2 + size/2),
c(lon_2 - size/2, lat_2 - size/2)
))))
# Set the number of mini rectangles
n <- 4
# Create random mini rectangles within the square
mini_rectangles_1 <- vector("list", n)
for (i in seq_len(n)) {
x1 <- runif(1, lon_1 - size/2, lon_1)
x2 <- runif(1, lon_1, lon_1 + size/2)
y1 <- runif(1, lat_1 - size/2, lat_1)
y2 <- runif(1, lat_1, lat_1 + size/2)
mini_rectangles_1[[i]] <- st_polygon(list(rbind(
c(x1, y1),
c(x2, y1),
c(x2,y2),
c(x1,y2),
c(x1,y1)
)))
}
mini_rectangles_1 <- st_sfc(mini_rectangles_1)
mini_rectangles_2 <- vector("list", n)
for (i in seq_len(n)) {
x1 <- runif(1, lon_2 - size/2, lon_2)
x2 <- runif(1, lon_2, lon_2 + size/2)
y1 <- runif(1,lat_2 - size/2,lat_2)
y2 <- runif(1,lat_2,lat_2 + size/2)
mini_rectangles_2[[i]] <- st_polygon(list(rbind(
c(x1,y1),
c(x2,y1),
c(x2,y2),
c(x1,y2),
c(x1,y1)
)))
}
mini_rectangles_2 <- st_sfc(mini_rectangles_2)
I then tried to visualize the results:
# Plot the squares and the mini rectangles
ggplot() +
geom_sf(data = st_sf(square_1), fill = "white", color = "black") +
geom_sf(data = st_sf(square_2), fill = "white", color = "black") +
geom_sf(data = st_sf(mini_rectangles_1), fill = "red", color = "red", alpha = 0.5) +
geom_sf(data = st_sf(mini_rectangles_2), fill = "blue", color = "blue", alpha = 0.5) +
coord_sf(xlim = c(min(lon_1 - size*0.6, lon_2 - size*0.6), max(lon_1 + size*0.6, lon_2 + size*0.6)), ylim = c(min(lat_1 - size*0.6, lat_2 - size*0.6), max(lat_1 + size*0.6, lat_2 + size*0.6)))
My Question: I am now interested in determining what percent of each red square is covered by each blue square and what percent of each blue square is covered by each red square
I tried to find some similar questions e.g. How to compute all pairwise interaction between polygons and the the percentage coverage in R with sf? - based on these references, I then tried to modify the code to suit my requirements:
# Create a 4x4 matrix to store coverage percentages
n = 4
coverage_matrix <- matrix(0,n,n)
# Calculate coverage percentages for each pair of red and blue rectangles
for (i in seq_len(n)) {
for (j in seq_len(n)) {
intersection_area <- st_area(st_intersection(mini_rectangles_1[[i]], mini_rectangles_2[[j]]))
red_area <- st_area(mini_rectangles_1[[i]])
coverage_matrix[i,j] <- 100 * intersection_area / red_area
}
}
# Print coverage matrix
print(coverage_matrix)
rownames(coverage_matrix) <- paste0("Red ", seq_len(n))
colnames(coverage_matrix) <- paste0("Blue ", seq_len(n))
# Print the modified coverage matrix
print(coverage_matrix)
The final result looks something like this:
Blue 1 Blue 2 Blue 3 Blue 4
Red 1 0.00000 0.00000 1.696167 16.371502
Red 2 0.00000 0.00000 23.741099 26.089504
Red 3 81.49752 19.38099 21.055805 47.402298
Red 4 28.27181 0.00000 0.000000 3.646122
Assuming I have done this correctly - I am trying to figure out a way to improve the speed of this code. In real life, I have multiple blue squares and multiple red squares (e.g. over 1000) and the code I have written is taking a very long time to run. As such, I am looking for ways to improve the efficiency and run-time of my code.
Can someone please show me how to do this?
References: