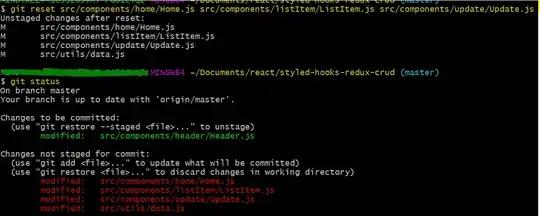
TL;DR: You can try the following:
library(terra)
library(purrr)
the_data <- read.csv('path/to/your/file.csv', header = FALSE,
blank.lines.skip = FALSE ## !
)
the_raster <- the_data |>
as.matrix() |>
(\(x) ifelse(x == '', NA, x))() |>
rast()
the_chunks <- patches(the_raster)
get_chunk_index <- function(m){
matrix(m, nrow(m), ncol(m), byrow = TRUE) |>
apply(MARGIN = 1 , FUN = \(row) paste0('chunk_',unique(row)[1]))
}
data_chunks <- the_data |> split(f = list(get_chunk_index(the_chunks)))
## grab activity data:
keep(data_chunks, ~ any(grepl('Activities', .x)))
how it works
- read in your data with your csv-reader of choice
- convert the resulting dataframe to a matrix (ensuring that empty cells of the originating spreadsheet are converted to NA)
- convert the matrix to a raster and use a package for image/spatial analysis (here: {terra}) to find patches of continguos non-empty cells (the analog to
CTRL+A in spreasheet software
- reconvert the raster of patches (=data chunks) to a matrix and extract the vector of patch numbers each row belongs to (or
NA for empty lines)
- split your initial data by this index vector, and extract/process the desired chunk from this list as desired.
- get data. Remember to set your csv-importer to keep blank lines (as those are used to distinguish between data blocks).
the_data <- read.csv('path/to/your/file.csv', header = FALSE,
blank.lines.skip = FALSE
)
- convert to matrix, define
NAs (here: empty strings), convert to spatial raster, inspect:
the_raster <- the_data |>
as.matrix() |>
(\(x) ifelse(x == '', NA, x))() |>
rast()
plot(the_raster)
- find continguous non-NA raster ranges (think
CTRL + A), inspect:
the_chunks <- patches(the_raster)
plot(the_chunks)
- helper function to extract the first non-NA row value (=chunk index) from a matrix:
get_chunk_index <- function(m){
matrix(m, nrow(m), ncol(m), byrow = TRUE) |>
apply(MARGIN = 1 , FUN = \(row) paste0('chunk_',unique(row)[1]))
}
- split initial dataframe by chunk index and get a list of data chunks:
data_chunks <- the_data |> split(f = list(get_chunk_index(the_chunks)))
data_chunks
# > data_chunks
$chunk_1
V1 V2 V3
1 Some junk Some junk Some junk
2 Some junk Some junk Some junk
3 Some junk Some junk Some junk
$chunk_2
V1 V2 V3
8 Activities
9 A1 A2 A3
10 2 1 3
11 3 4 6
# ...
- proceed with the desired chunks. Example: pick any chunk containing "Activity":
library(purrr) ## for convenient list operations
keep(data_chunks, ~ any(grepl('Activities', .x)))
$chunk_2
V1 V2 V3
8 Activities
9 A1 A2 A3
10 2 1 3
11 3 4 6
I used this example structure: