In R, there are many factors which can affect performance. In order for others to provide 'good' answers, it would be helpful if you could edit your question to include your current runtime, the sort of speed-up you expect, and a minimal reproducible example (alternatively, describe any differences between your actual data and my example below, e.g. how long is your target_codes vector, what are the dimensions of your dataframes, etc). Also, how many dataframes are you applying this to, and have you looked at other alternatives, e.g. data.table, or other languages, e.g. awk?
Your code didn't run 'as is' on my example dataset, so I ran an altered version. I also tested PCRE regex (perl = TRUE in the grep) which can improve speed for reasons:
library(tidyverse)
match_targets_dplyr <- function(df, target_codes){
pattern = paste0("^", target_codes, collapse = "|")
df <- filter(df, if_any(everything(), ~grepl(pattern, .x)))
return(df)
}
match_targets_dplyr_perl <- function(df, target_codes){
pattern = paste0("^", target_codes, collapse = "|")
df <- filter(df, if_any(everything(), ~grepl(pattern, .x, perl = TRUE)))
return(df)
}
match_targets_base <- function(df, target_codes){
pattern = paste0("^", target_codes, collapse = "|")
df <- df[rowSums(apply(df, 2, \(x) grepl(pattern, x))) > 0, , drop = FALSE]
return(df)
}
match_targets_base_perl <- function(df, target_codes){
pattern = paste0("^", target_codes, collapse = "|")
df <- df[rowSums(apply(df, 2, \(x) grepl(pattern, x, perl = TRUE))) > 0, , drop = FALSE]
return(df)
}
# grab some data
df <- read_csv("https://raw.github.com/VladAluas/Text_Analysis/master/Datasets/Text_review.csv")
#> Rows: 433 Columns: 3
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (3): Model, Segment, Text
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
df_large <- df
# the "Text" field has some non-standard characters; remove them
df_large$Text <- iconv(df_large$Text, from = "UTF-8", to = "ASCII", sub = "")
# select some target codes
target_codes = c("Perform", "Ca", "Net")
# check you get the same answer from all of the functions
df2 <- match_targets_dplyr(df_large, target_codes)
df3 <- match_targets_dplyr_perl(df_large, target_codes)
df4 <- match_targets_base(df_large, target_codes)
df5 <- match_targets_base_perl(df_large, target_codes)
all.equal(df2, df3, df4, df5, check.attributes = FALSE)
#> [1] TRUE
# increase the size of the data to ~28mil rows
repeat{df_large <- df_large %>% bind_rows(df_large); if(NROW(df_large) > 2.8e7) {break}}
# benchmark the functions on the 28mil rows
library(microbenchmark)
res <- microbenchmark(match_targets_dplyr(df_large, target_codes),
match_targets_dplyr_perl(df_large, target_codes),
match_targets_base(df_large, target_codes),
match_targets_base_perl(df_large, target_codes),
times = 2)
autoplot(res)
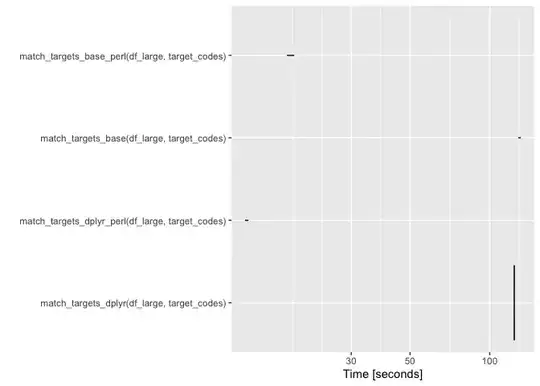
Created on 2023-06-02 with reprex v2.0.2
It's hard to see what's going on in the reprex image; here it is with the points highlighted:
TL,DR: dplyr is a bit faster, and PCRE was much faster using this example dataset.

