I am using R Markdown to graph a volcano plot of genes using the Seurat and EnhancedVolcano packages.
This is my code below. The 'vgm' is a Seurat Object:
#using Seurat to obtain differentially expressed genes between two groups, labelled 4 vs 6
DEgenes<- Seurat::FindMarkers(vgm[[2]], group.by = "Group", ident.1="4",ident.2="6", min.pct=0.25)
#results stored in a dataframe
head(DEgenes)
Results of the head() function in the code above produces this output: Output dataframe
Next, I proceed to run the following codes:
#installing and loading EnhancedVolcano
devtools::install_github('kevinblighe/EnhancedVolcano')
library(EnhancedVolcano)
#plotting volcano plot between group 4 (called InEx) vs group 6 (called ReA SF)
#For InEx vs ReA SF
EnhancedVolcano(DEgenes,
lab = rownames(DEgenes),
x = 'avg_log2FC',
y = 'p_val_adj',
ylim = c(0,450),
title = 'InEx vs ReA SF',
xlab = bquote(~Log[1]~ '(Fold Change)'),
ylab = bquote(~-Log[10]~ '(Adj. p)'),
axisLabSize = 12,
FCcutoff = 1.493,
pCutoff = 10e-40,
pointSize = 1,
drawConnectors = TRUE,
widthConnectors = 0.75,
colAlpha = 1,
max.overlaps = 45)
When I run the code chunk above for my figure, it is displayed within the R Markdown output normally as seen below. As seen in this picture, the labels of certain genes are intact and visible:
When I want to see it in a new window, I press this button here (circled in purple):
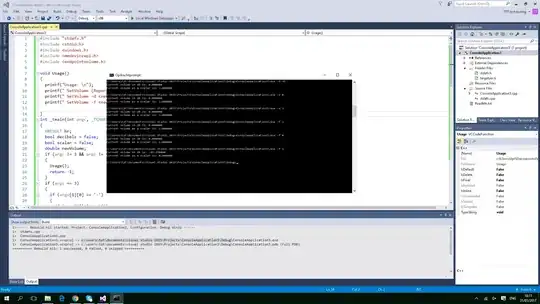
However, when this new window is opened, the gene labels are not visible anymore like it was before:

I am not sure why the entirety of the plot is not showing up in the new window. Any ideas on having the gene labels visible in the new window would be greatly welcome!
Thanks in advance! I edited this question to make it as reproducible as possible.