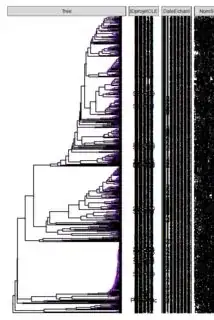
I managed to plot a phylogenetic tree using ggtree and pdf() but cant break it down to fit on multiple pages to make it readable for users.
You can make a tree following this link
Heres the code for the tree and its output :
make_tree_plot <- function(phy_tree, tip_label){
p <- ggtree(phy_tree) + geom_nodepoint(color='purple', size=2, alpha=0.2) + geom_tippoint()
...# a lot of code to create facet_plots to have all the variables included in the panel. We end up with the final form p11 :
assign(p11, facet_plot(z11), panel=colnames(info_df[i]), geom=geom_text,
aes(x=0, label=unlist(info_df[11])), data=info_df))
gt <- ggplot_gtable(ggplot_build(p11))
grid.draw(gt)
}
# Master
pdf("plots2.pdf", width=35, height=45, paper='a4r')
make_tree_plot(tree, tips_info)
dev.off()
We get this CONDENSED tree in one-page pdf.
This is the expected output.
Considering the nature of the plot, I can't simply divide the dataset and create multiple trees. Only one tree can be created and then divided in multiple pages.
I am coding in VS code and have tried .rmd files for printing but it wasn't the ideal for me. I managed to print matrixes over multiple pages with pdf(), but for this phylo tree im honestly facing a wall.