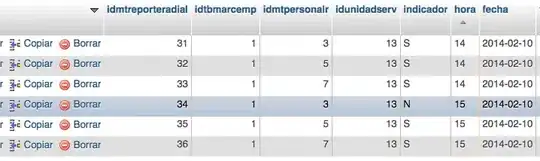
I have two arrays, x and y, which determine the position and a third one, z, which provides the value of the radiation at such position.
I want to calculate and plot the HDI of 90% over the plot of radiation to be able to say that 90% of the radiation can be found in such region.
If I were to normalize z,the HDI for 2D radiation data means the smallest area in which one can integrate the normalized distribution function and get the desired level, in my case, 0.9 or 90%, being the integral between -inf and inf equal to 1.0.
This problem looks isomorphic to calculate the HDI of a probability distribution so I think that it can be done, but I just do not know how to proceed any further.
Edit to make it clearer: This snipped generates data (x,y, and z) which are similar to the topology of my own, so I am using it as a proxy:
import numpy as np
## Spatial data, with randomness to show that it is not in a rectangular grid
x = np.arange(-2,2, step=0.2)
x+= 0.1*np.random.rand(len(x))
y = np.arange(-2,2, step=0.2)
y+= 0.1*np.random.rand(len(y))
z = np.zeros((len(x), len(y)))
for i, ix in enumerate(x):
for k, iy in enumerate(y):
tmp = np.cos(ix)*np.sin(iy)
if tmp>=0:
z[i,k] = tmp
z=z/np.sum(z)
This is a crude sketch of my expectations: The data are mostly accumulated on the left, as shown by the contour plots, and the red line represents HDI of .9, where 90% of the integral of data is enclosed (the line is purposely not following one of the isolines, because that needn't to be the case)
The bold black line labelled 94% HDI is an example of what I want, but I want it in 2D:
I cannot find the way to use az.hdi, or az.plot_kde.
I have tried to feed the data as observable to a PYMC model, so that the MCMC sampling could translate values to density of points (which is what az.hdi and az.plot_kde want), but I haven't been able to do it. A very crude approach would be to generate artificial points around each of my own proportional to the value of radiation at that point. I think there must be a more elegant and efficient way to solve this.
Getting the indices of the points inside the HDI could be an alternative solution, because calculating the convex hull I could still get the HDI.
Any idea about how to proceed is welcome.