I am facing issues with stat_compare_means, geom_pwc, and stat_pvalue_manual. The issues are as follows:
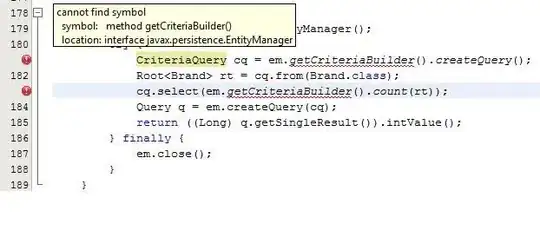
stat_compare_meansactually works decently for the most part, except for the fact that I cannot seem to get rid of the brackets of NS significance.hide.nsonly gets rid of the label itself but not the brackets, and I cannot seem to get those brackets to not appear.With both
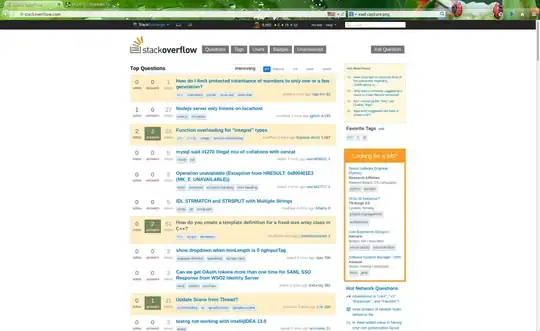
geom_pwcandstat_pvalue_manual, I read thathide.nsalso hides brackets. However, when I use either of these functions withhide.ns=TRUE, all of my significances are removed (despite not all of them being ns).
My data is a df with three relevant variables: willingness (y-axis), gender (x-axis), and risk (facet). My plot is a boxplot. There are three facets (risk: 0.1%, 2%, and 10%), four gender categories (men, women, non-binary, and general), and willingness is a continuous 0-100 scale. The "general" gender is just the combined data of the other three gender categories, to observe the general trend non-gender-specific, and should be left out of the statistical comparisons.
My code is as follows:
ggboxplot(combined_data,x = "gender", y = "willingness", color = "black", fill="gender", palette = c("coral4","#BBD1EA","#FFBB99","#FFF6BD"), facet.by = "Risk", short.panel.labs = FALSE, outlier.shape = NA) +
geom_point(aes(x=gender,y=willingness,fill=gender),position=position_jitterdodge(),alpha=0.3,size=1,shape=20) +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) +
theme(legend.position="none", axis.text.x=element_blank(),axis.ticks.x=element_blank(),axis.title.x = element_blank()) +
stat_compare_means(comparisons = my_comparisons, label="p.signif", hide.ns = TRUE)
Where, in stat_compare_means, "my_comparisons" is:
my_comparisons <- list(c("Men", "Women"), c("Men", "Non-binary"), c("Women", "Non-binary"))
This produces the first image attached below.
When I try to use geom_pwc or stat_pvalue_manual, they both produce the same figure. Note: this one does not seem to allow me to specify the comparisons, so "general" is included here. If anyone knows how to specify comparisons, that would be great. The code is:
ggboxplot(combined_data,x = "gender", y = "willingness", color = "black", fill="gender", palette = c("coral4","#BBD1EA","#FFBB99","#FFF6BD"), facet.by = "Risk", short.panel.labs = FALSE, outlier.shape = NA) +
geom_point(aes(x=gender,y=willingness,fill=gender),position=position_jitterdodge(),alpha=0.3,size=1,shape=20) +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) +
theme(legend.position="none", axis.text.x=element_blank(),axis.ticks.x=element_blank(),axis.title.x = element_blank()) +
geom_pwc(group.by="x.var",label="p.signif",hide.ns=TRUE)
And the figure is found below.
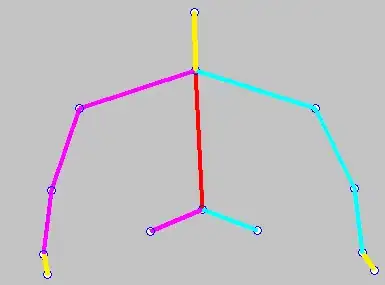
And this is what it looks like with hide.ns=false, noting the present significance levels:
Using stat_pvalue_manual produces the same result as geom_pwc, noting the following code:
stat.test <- compare_means(willingness ~ gender, data = combined_data,
group.by = "Risk")
ggboxplot(.......same as above) +
stat_pvalue_manual(stat.test, label = "p.signif", hide.ns = TRUE)
So if anyone can help with either:
- Removing ns brackets with stat_compare_means, or
- Specifying comparisons and fixing the
hide.nsissue withgeom_pwcorstat_pvalue_manual
I would really appreciate it. Thanks!