I am trying to create a Sankey or Alluvial plot using the ggplot2 library in R to visualize the flow of nodes based on the provided CSV data. The data includes columns for 'x', 'node', 'next_x', and 'next_node'. I want to create a plot where the flow is determined by the 'node' and 'next_node' columns. Additionally, I want to exclude any flows where 'next_x' is "NA".
Here's a simplified version of the CSV data I'm working with:
x node next_x next_node
Homo_sapiens SLC35A1 Mus_musculus SLC35A1
Homo_sapiens RARS2 Mus_musculus RARS2
Homo_sapiens ORC3 Mus_musculus ORC3
Homo_sapiens AKIRIN2 Mus_musculus AKIRIN2
Homo_sapiens SPACA1 Mus_musculus SPACA1
Homo_sapiens CNR1 Mus_musculus CNR1
Homo_sapiens RNGTT Mus_musculus RNGTT
Homo_sapiens PNRC1 Mus_musculus PNRC1
Homo_sapiens PM20D2 Mus_musculus PM20D2
Homo_sapiens SRSF12 Mus_musculus SRSF12
Homo_sapiens GABRR1 Mus_musculus GABRR1
Mus_musculus GABRR1 Rattus_norvegicus GABRR1
Mus_musculus PM20D2 Rattus_norvegicus PM20D2
Mus_musculus SRSF12 Rattus_norvegicus SRSF12
Mus_musculus PNRC1 Rattus_norvegicus PNRC1
Mus_musculus RNGTT Rattus_norvegicus RNGTT
Mus_musculus CNR1 Rattus_norvegicus CNR1
Mus_musculus SPACA1 Rattus_norvegicus SPACA1
Mus_musculus AKIRIN2 Rattus_norvegicus AKIRIN2
Mus_musculus ORC3 Rattus_norvegicus ORC3
Mus_musculus RARS2 Rattus_norvegicus RARS2
Mus_musculus SLC35A1 Rattus_norvegicus SLC35A1
Rattus_norvegicus GABRR1 Canis_lupus_familiaris GABRR1
Rattus_norvegicus PM20D2 Canis_lupus_familiaris PM20D2
Rattus_norvegicus SRSF12 Canis_lupus_familiaris SRSF12
Rattus_norvegicus PNRC1 Canis_lupus_familiaris PNRC1
Rattus_norvegicus RNGTT Canis_lupus_familiaris RNGTT
Rattus_norvegicus CNR1 Canis_lupus_familiaris CNR1
Rattus_norvegicus SPACA1 Canis_lupus_familiaris SPACA1
Rattus_norvegicus AKIRIN2 Canis_lupus_familiaris AKIRIN2
Rattus_norvegicus ORC3 Canis_lupus_familiaris ORC3
Rattus_norvegicus RARS2 Canis_lupus_familiaris RARS2
Rattus_norvegicus SLC35A1 Canis_lupus_familiaris SLC35A1
Canis_lupus_familiaris SLC35A1 Monodelphis_domestica SLC35A1
Canis_lupus_familiaris RARS2 Monodelphis_domestica RARS2
Canis_lupus_familiaris ORC3 Monodelphis_domestica ORC3
Canis_lupus_familiaris AKIRIN2 Monodelphis_domestica AKIRIN2
Canis_lupus_familiaris SPACA1 Monodelphis_domestica SPACA1
Canis_lupus_familiaris CNR1 Monodelphis_domestica CNR1
Canis_lupus_familiaris RNGTT Monodelphis_domestica RNGTT
Canis_lupus_familiaris PNRC1 Monodelphis_domestica PNRC1
Canis_lupus_familiaris SRSF12 Monodelphis_domestica SRSF12
Canis_lupus_familiaris PM20D2 Monodelphis_domestica PM20D2
Canis_lupus_familiaris GABRR1 Monodelphis_domestica GABRR1
Monodelphis_domestica SLC35A1 Ornithorhynchus_anatinus SLC35A1
Monodelphis_domestica RARS2 Ornithorhynchus_anatinus RARS2
Monodelphis_domestica ORC3 Ornithorhynchus_anatinus ORC3
Monodelphis_domestica AKIRIN2 Ornithorhynchus_anatinus AKIRIN2
Monodelphis_domestica SPACA1 Ornithorhynchus_anatinus SPACA1
Monodelphis_domestica CNR1 Ornithorhynchus_anatinus CNR1
Monodelphis_domestica RNGTT Ornithorhynchus_anatinus RNGTT
Monodelphis_domestica PNRC1 Ornithorhynchus_anatinus PNRC1
Monodelphis_domestica SRSF12 NA NA
Monodelphis_domestica PM20D2 Ornithorhynchus_anatinus PM20D2
Monodelphis_domestica GABRR1 NA NA
Ornithorhynchus_anatinus SLC35A1 Gallus_gallus SLC35A1
Ornithorhynchus_anatinus RARS2 Gallus_gallus RARS2
Ornithorhynchus_anatinus ORC3 Gallus_gallus ORC3
Ornithorhynchus_anatinus AKIRIN2 Gallus_gallus AKIRIN2
Ornithorhynchus_anatinus SPACA1 Gallus_gallus SPACA1
Ornithorhynchus_anatinus CNR1 Gallus_gallus CNR1
Ornithorhynchus_anatinus RNGTT Gallus_gallus RNGTT
Ornithorhynchus_anatinus PNRC1 Gallus_gallus PNRC1
Ornithorhynchus_anatinus PM20D2 Gallus_gallus PM20D2
Ornithorhynchus_anatinus LOC100076186 NA NA
Ornithorhynchus_anatinus LOC114805750 NA NA
Gallus_gallus PM20D2 Taeniopygia_guttata PM20D2
Gallus_gallus PNRC1 Taeniopygia_guttata PNRC1
Gallus_gallus BORCS6 Taeniopygia_guttata BORCS6
Gallus_gallus RNGTT Taeniopygia_guttata RNGTT
Gallus_gallus LOC101749895 NA NA
Gallus_gallus CNR1 Taeniopygia_guttata CNR1
Gallus_gallus SPACA1 NA NA
Gallus_gallus AKIRIN2 Taeniopygia_guttata AKIRIN2
Gallus_gallus ORC3 Taeniopygia_guttata ORC3
Gallus_gallus RARS2 Taeniopygia_guttata RARS2
Gallus_gallus SLC35A1 Taeniopygia_guttata SLC35A1
Taeniopygia_guttata CFAP206 NA NA
Taeniopygia_guttata SLC35A1 Chelonia_mydas SLC35A1
Taeniopygia_guttata RARS2 Chelonia_mydas RARS2
Taeniopygia_guttata ORC3 Chelonia_mydas ORC3
Taeniopygia_guttata AKIRIN2 Chelonia_mydas AKIRIN2
Taeniopygia_guttata CNR1 Chelonia_mydas CNR1
Taeniopygia_guttata RNGTT Chelonia_mydas RNGTT
Taeniopygia_guttata BORCS6 NA NA
Taeniopygia_guttata PNRC1 Chelonia_mydas PNRC1
Taeniopygia_guttata PM20D2 Chelonia_mydas PM20D2
Taeniopygia_guttata GABRR1 Chelonia_mydas GABRR1
Chelonia_mydas SLC35A1 Anolis_carolinensis SLC35A1
Chelonia_mydas RARS2 Anolis_carolinensis RARS2
Chelonia_mydas ORC3 Anolis_carolinensis ORC3
Chelonia_mydas AKIRIN2 Anolis_carolinensis AKIRIN2
Chelonia_mydas SPACA1 Anolis_carolinensis SPACA1
Chelonia_mydas CNR1 Anolis_carolinensis CNR1
Chelonia_mydas RNGTT Anolis_carolinensis RNGTT
Chelonia_mydas LOC102938330 NA NA
Chelonia_mydas PNRC1 Anolis_carolinensis PNRC1
Chelonia_mydas PM20D2 Anolis_carolinensis PM20D2
Chelonia_mydas GABRR1 NA NA
Anolis_carolinensis PM20D2 NA NA
Anolis_carolinensis SRSF12 NA NA
Anolis_carolinensis PNRC1 NA NA
Anolis_carolinensis RNGTT NA NA
Anolis_carolinensis LOC107982676 NA NA
Anolis_carolinensis CNR1 NA NA
Anolis_carolinensis SPACA1 NA NA
Anolis_carolinensis AKIRIN2 NA NA
Anolis_carolinensis ORC3 NA NA
Anolis_carolinensis RARS2 NA NA
Anolis_carolinensis SLC35A1 NA NA
Xenopus_laevis GABRR2.S NA NA
Xenopus_laevis GABRR1.S NA NA
Xenopus_laevis PM20D2.S NA NA
Xenopus_laevis LOC108717975 NA NA
Xenopus_laevis RNGTT.S NA NA
Xenopus_laevis CNR1.S NA NA
Xenopus_laevis AKIRIN2.S NA NA
Xenopus_laevis ORC3.S NA NA
Xenopus_laevis RARS2.S NA NA
Xenopus_laevis SLC35A1.S NA NA
Xenopus_laevis LOC108717977 NA NA
Latimeria_chalumnae DDX24 NA NA
Latimeria_chalumnae PPP4R4 NA NA
Latimeria_chalumnae SERPINA10B NA NA
Latimeria_chalumnae ARRDC3A NA NA
Latimeria_chalumnae LOC102360869 NA NA
Latimeria_chalumnae CNR1 Protopterus_annectens CNR1
Latimeria_chalumnae SPACA1 NA NA
Latimeria_chalumnae AKIRIN2 NA NA
Latimeria_chalumnae ORC3 NA NA
Latimeria_chalumnae RARS2 NA NA
Latimeria_chalumnae LOC102362557 NA NA
Protopterus_annectens LOC122794922 NA NA
Protopterus_annectens LOC122794923 NA NA
Protopterus_annectens LOC122794924 NA NA
Protopterus_annectens FBXL5 NA NA
Protopterus_annectens CC2D2A NA NA
Protopterus_annectens CNR1 Danio_rerio CNR1
Protopterus_annectens CPEB2 NA NA
Protopterus_annectens BOD1L1 NA NA
Protopterus_annectens C1QTNF7 NA NA
Protopterus_annectens NKX3-2 NA NA
Protopterus_annectens RAB28 NA NA
Danio_rerio MYO6A NA NA
Danio_rerio LOC569340 NA NA
Danio_rerio MEI4 NA NA
Danio_rerio NT5E NA NA
Danio_rerio SNX14 NA NA
Danio_rerio CNR1 Oreochromis_niloticus CNR1
Danio_rerio RNGTT Oreochromis_niloticus RNGTT
Danio_rerio PNRC1 NA NA
Danio_rerio GABRR1 NA NA
Danio_rerio GABRR2B NA NA
Danio_rerio UBE2J1 NA NA
Oreochromis_niloticus SI:DKEY-174M14.3 NA NA
Oreochromis_niloticus RDH14B NA NA
Oreochromis_niloticus LOC102078481 NA NA
Oreochromis_niloticus RNGTT Scyliorhinus_canicula RNGTT
Oreochromis_niloticus LOC112842425 NA NA
Oreochromis_niloticus CNR1 Scyliorhinus_canicula CNR1
Oreochromis_niloticus AKIRIN2 Scyliorhinus_canicula AKIRIN2
Oreochromis_niloticus RARS2 Scyliorhinus_canicula RARS2
Oreochromis_niloticus SLC35A1 Scyliorhinus_canicula SLC35A1
Oreochromis_niloticus LOC100692709 NA NA
Oreochromis_niloticus LOC102081816 NA NA
Scyliorhinus_canicula SLC35A1 Petromyzon_marinus SLC35A1
Scyliorhinus_canicula RARS2 Petromyzon_marinus RARS2
Scyliorhinus_canicula ORC3 Petromyzon_marinus ORC3
Scyliorhinus_canicula AKIRIN2 Petromyzon_marinus AKIRIN2
Scyliorhinus_canicula LOC119967921 NA NA
Scyliorhinus_canicula CNR1 Petromyzon_marinus CNR1
Scyliorhinus_canicula RNGTT Petromyzon_marinus RNGTT
Scyliorhinus_canicula LOC119967175 NA NA
Scyliorhinus_canicula PNRC1 NA NA
Scyliorhinus_canicula LOC119967178 NA NA
Scyliorhinus_canicula LOC119967180 NA NA
Petromyzon_marinus LOC116953416 NA NA
Petromyzon_marinus LOC116953419 NA NA
Petromyzon_marinus CEP162 NA NA
Petromyzon_marinus FBXL22 NA NA
Petromyzon_marinus RNGTT NA NA
Petromyzon_marinus CNR1 NA NA
Petromyzon_marinus AKIRIN2 NA NA
Petromyzon_marinus ORC3 NA NA
Petromyzon_marinus RARS2 NA NA
Petromyzon_marinus SLC35A1 NA NA
Petromyzon_marinus RHBDL2 NA NA
I'm using the ggplot2 library to create the plot, and I've tried the following script:
library(ggplot2)
pl <- ggplot(data, aes(x = x, node = node, next_node = next_node, next_x = next_x, fill = factor(node), label = node)) +
geom_sankey(flow.alpha = 0.5,
node.color = "black",
show.legend = FALSE,
na.rm = TRUE) +
geom_sankey_label(size = 3, color = "black", fill="white", hjust = 0.5) +
theme_bw() +
theme(legend.position = "none") +
theme(axis.title = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank(),
panel.grid = element_blank()) +
scale_fill_viridis_d(option = "inferno") +
labs(title = "Sankey diagram using ggplot",
fill = "Nodes")
However, when I run this script, I'm encountering the following warning messages:
Warning messages:
1: There was 1 warning in `dplyr::mutate()`.
ℹ In argument: `dplyr::across(c(x, next_x), ~as.numeric(.), .names = ("n_{.col}"))`.
Caused by warning:
! NAs introduced by coercion
2: There was 1 warning in `dplyr::mutate()`.
ℹ In argument: `dplyr::across(c(x, next_x), ~as.numeric(.), .names = ("n_{.col}"))`.
Caused by warning:
! NAs introduced by coercion
3: There was 1 warning in `dplyr::mutate()`.
ℹ In argument: `dplyr::across(c(x, next_x), ~as.numeric(.), .names = ("n_{.col}"))`.
Caused by warning:
! NAs introduced by coercion
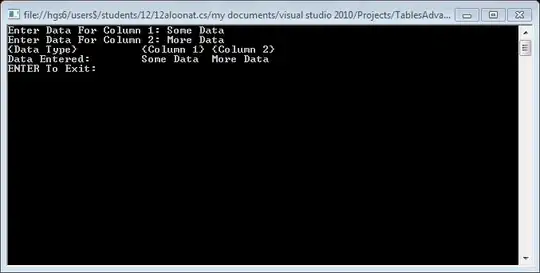
I also get an incomplete plot:
Incomplete Sankey plot without flow
I'm seeking guidance on how to address this issue and successfully create the desired Sankey or Alluvial plot using ggplot2. Specifically, I want to achieve the following:
- Create a plot where the flow is based on 'node' and 'next_node'.
- Exclude flows where 'next_x' is "NA".
- Avoid the warning messages related to dplyr::mutate() and NAs.
Any assistance or insights into solving this problem would be greatly appreciated. Thank you in advance!
Edit:
This is my raw dataset of gene neighbors:
species gene start stop orientation
Homo_sapiens SLC35A1 1 2 1
Homo_sapiens RARS2 2 3 -1
Homo_sapiens ORC3 3 4 1
Homo_sapiens AKIRIN2 4 5 -1
Homo_sapiens SPACA1 5 6 1
Homo_sapiens CNR1 6 7 -1
Homo_sapiens RNGTT 7 8 -1
Homo_sapiens PNRC1 8 9 1
Homo_sapiens PM20D2 9 10 1
Homo_sapiens SRSF12 10 11 -1
Homo_sapiens GABRR1 11 12 -1
Mus_musculus GABRR1 1 2 1
Mus_musculus PM20D2 2 3 -1
Mus_musculus SRSF12 3 4 1
Mus_musculus PNRC1 4 5 -1
Mus_musculus RNGTT 5 6 1
Mus_musculus CNR1 6 7 1
Mus_musculus SPACA1 7 8 -1
Mus_musculus AKIRIN2 8 9 1
Mus_musculus ORC3 9 10 -1
Mus_musculus RARS2 10 11 1
Mus_musculus SLC35A1 11 12 -1
Rattus_norvegicus GABRR1 1 2 1
Rattus_norvegicus PM20D2 2 3 -1
Rattus_norvegicus SRSF12 3 4 1
Rattus_norvegicus PNRC1 4 5 -1
Rattus_norvegicus RNGTT 5 6 1
Rattus_norvegicus CNR1 6 7 1
Rattus_norvegicus SPACA1 7 8 -1
Rattus_norvegicus AKIRIN2 8 9 1
Rattus_norvegicus ORC3 9 10 -1
Rattus_norvegicus RARS2 10 11 1
Rattus_norvegicus SLC35A1 11 12 -1
Canis_lupus_familiaris SLC35A1 1 2 1
Canis_lupus_familiaris RARS2 2 3 -1
Canis_lupus_familiaris ORC3 3 4 1
Canis_lupus_familiaris AKIRIN2 4 5 -1
Canis_lupus_familiaris SPACA1 5 6 1
Canis_lupus_familiaris CNR1 6 7 -1
Canis_lupus_familiaris RNGTT 7 8 -1
Canis_lupus_familiaris PNRC1 8 9 1
Canis_lupus_familiaris SRSF12 9 10 -1
Canis_lupus_familiaris PM20D2 10 11 1
Canis_lupus_familiaris GABRR1 11 12 -1
Monodelphis_domestica SLC35A1 1 2 1
Monodelphis_domestica RARS2 2 3 -1
Monodelphis_domestica ORC3 3 4 1
Monodelphis_domestica AKIRIN2 4 5 -1
Monodelphis_domestica SPACA1 5 6 1
Monodelphis_domestica CNR1 6 7 -1
Monodelphis_domestica RNGTT 7 8 -1
Monodelphis_domestica PNRC1 8 9 1
Monodelphis_domestica SRSF12 9 10 -1
Monodelphis_domestica PM20D2 10 11 1
Monodelphis_domestica GABRR1 11 12 -1
Ornithorhynchus_anatinus SLC35A1 1 2 1
Ornithorhynchus_anatinus RARS2 2 3 -1
Ornithorhynchus_anatinus ORC3 3 4 1
Ornithorhynchus_anatinus AKIRIN2 4 5 -1
Ornithorhynchus_anatinus SPACA1 5 6 1
Ornithorhynchus_anatinus CNR1 6 7 -1
Ornithorhynchus_anatinus RNGTT 7 8 -1
Ornithorhynchus_anatinus PNRC1 8 9 1
Ornithorhynchus_anatinus PM20D2 9 10 1
Ornithorhynchus_anatinus LOC100076186 10 11 -1
Ornithorhynchus_anatinus LOC114805750 11 12 1
Gallus_gallus PM20D2 1 2 -1
Gallus_gallus PNRC1 2 3 -1
Gallus_gallus BORCS6 3 4 1
Gallus_gallus RNGTT 4 5 1
Gallus_gallus LOC101749895 5 6 1
Gallus_gallus CNR1 6 7 1
Gallus_gallus SPACA1 7 8 -1
Gallus_gallus AKIRIN2 8 9 1
Gallus_gallus ORC3 9 10 -1
Gallus_gallus RARS2 10 11 1
Gallus_gallus SLC35A1 11 12 -1
Taeniopygia_guttata CFAP206 1 2 1
Taeniopygia_guttata SLC35A1 2 3 1
Taeniopygia_guttata RARS2 3 4 -1
Taeniopygia_guttata ORC3 4 5 1
Taeniopygia_guttata AKIRIN2 5 6 -1
Taeniopygia_guttata CNR1 6 7 -1
Taeniopygia_guttata RNGTT 7 8 -1
Taeniopygia_guttata BORCS6 8 9 -1
Taeniopygia_guttata PNRC1 9 10 1
Taeniopygia_guttata PM20D2 10 11 1
Taeniopygia_guttata GABRR1 11 12 -1
Chelonia_mydas SLC35A1 1 2 1
Chelonia_mydas RARS2 2 3 -1
Chelonia_mydas ORC3 3 4 1
Chelonia_mydas AKIRIN2 4 5 -1
Chelonia_mydas SPACA1 5 6 1
Chelonia_mydas CNR1 6 7 -1
Chelonia_mydas RNGTT 7 8 -1
Chelonia_mydas LOC102938330 8 9 -1
Chelonia_mydas PNRC1 9 10 1
Chelonia_mydas PM20D2 10 11 1
Chelonia_mydas GABRR1 11 12 -1
Anolis_carolinensis PM20D2 1 2 -1
Anolis_carolinensis SRSF12 2 3 1
Anolis_carolinensis PNRC1 3 4 -1
Anolis_carolinensis RNGTT 4 5 1
Anolis_carolinensis LOC107982676 5 6 -1
Anolis_carolinensis CNR1 6 7 1
Anolis_carolinensis SPACA1 7 8 -1
Anolis_carolinensis AKIRIN2 8 9 1
Anolis_carolinensis ORC3 9 10 -1
Anolis_carolinensis RARS2 10 11 1
Anolis_carolinensis SLC35A1 11 12 -1
Xenopus_laevis GABRR2.S 1 2 1
Xenopus_laevis GABRR1.S 2 3 1
Xenopus_laevis PM20D2.S 3 4 -1
Xenopus_laevis LOC108717975 4 5 1
Xenopus_laevis RNGTT.S 5 6 1
Xenopus_laevis CNR1.S 6 7 1
Xenopus_laevis AKIRIN2.S 7 8 1
Xenopus_laevis ORC3.S 8 9 -1
Xenopus_laevis RARS2.S 9 10 1
Xenopus_laevis SLC35A1.S 10 11 -1
Xenopus_laevis LOC108717977 11 12 1
Latimeria_chalumnae DDX24 1 2 -1
Latimeria_chalumnae PPP4R4 2 3 1
Latimeria_chalumnae SERPINA10B 3 4 -1
Latimeria_chalumnae ARRDC3A 4 5 1
Latimeria_chalumnae LOC102360869 5 6 -1
Latimeria_chalumnae CNR1 6 7 1
Latimeria_chalumnae SPACA1 7 8 -1
Latimeria_chalumnae AKIRIN2 8 9 1
Latimeria_chalumnae ORC3 9 10 -1
Latimeria_chalumnae RARS2 10 11 1
Latimeria_chalumnae LOC102362557 11 12 1
Protopterus_annectens LOC122794922 1 2 1
Protopterus_annectens LOC122794923 2 3 1
Protopterus_annectens LOC122794924 3 4 1
Protopterus_annectens FBXL5 4 5 1
Protopterus_annectens CC2D2A 5 6 -1
Protopterus_annectens CNR1 6 7 1
Protopterus_annectens CPEB2 7 8 -1
Protopterus_annectens BOD1L1 8 9 -1
Protopterus_annectens C1QTNF7 9 10 -1
Protopterus_annectens NKX3-2 10 11 1
Protopterus_annectens RAB28 11 12 1
Danio_rerio MYO6A 1 2 1
Danio_rerio LOC569340 2 3 -1
Danio_rerio MEI4 3 4 1
Danio_rerio NT5E 4 5 1
Danio_rerio SNX14 5 6 -1
Danio_rerio CNR1 6 7 -1
Danio_rerio RNGTT 7 8 -1
Danio_rerio PNRC1 8 9 1
Danio_rerio GABRR1 9 10 -1
Danio_rerio GABRR2B 10 11 -1
Danio_rerio UBE2J1 11 12 -1
Oreochromis_niloticus SI:DKEY-174M14.3 1 2 1
Oreochromis_niloticus RDH14B 2 3 -1
Oreochromis_niloticus LOC102078481 3 4 1
Oreochromis_niloticus RNGTT 4 5 1
Oreochromis_niloticus LOC112842425 5 6 -1
Oreochromis_niloticus CNR1 6 7 1
Oreochromis_niloticus AKIRIN2 7 8 1
Oreochromis_niloticus RARS2 8 9 1
Oreochromis_niloticus SLC35A1 9 10 -1
Oreochromis_niloticus LOC100692709 10 11 -1
Oreochromis_niloticus LOC102081816 11 12 1
Scyliorhinus_canicula SLC35A1 1 2 1
Scyliorhinus_canicula RARS2 2 3 -1
Scyliorhinus_canicula ORC3 3 4 1
Scyliorhinus_canicula AKIRIN2 4 5 -1
Scyliorhinus_canicula LOC119967921 5 6 1
Scyliorhinus_canicula CNR1 6 7 -1
Scyliorhinus_canicula RNGTT 7 8 -1
Scyliorhinus_canicula LOC119967175 8 9 -1
Scyliorhinus_canicula PNRC1 9 10 1
Scyliorhinus_canicula LOC119967178 10 11 1
Scyliorhinus_canicula LOC119967180 11 12 -1
Petromyzon_marinus LOC116953416 1 2 -1
Petromyzon_marinus LOC116953419 2 3 -1
Petromyzon_marinus CEP162 3 4 1
Petromyzon_marinus FBXL22 4 5 -1
Petromyzon_marinus RNGTT 5 6 1
Petromyzon_marinus CNR1 6 7 1
Petromyzon_marinus AKIRIN2 7 8 1
Petromyzon_marinus ORC3 8 9 -1
Petromyzon_marinus RARS2 9 10 1
Petromyzon_marinus SLC35A1 10 11 -1
Petromyzon_marinus RHBDL2 11 12 1
Edit 2:
I've managed to get few flows connected but it is still incorrect. The problem is probably with the order of the rows. Can somebody please suggest something?