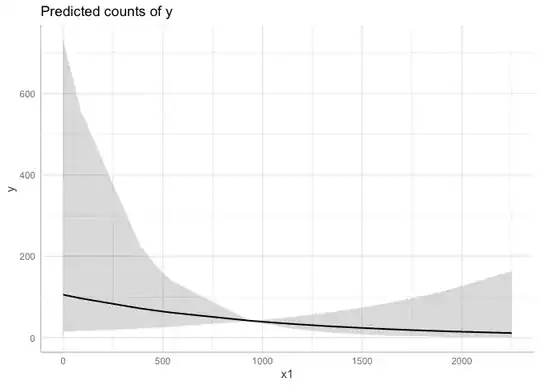
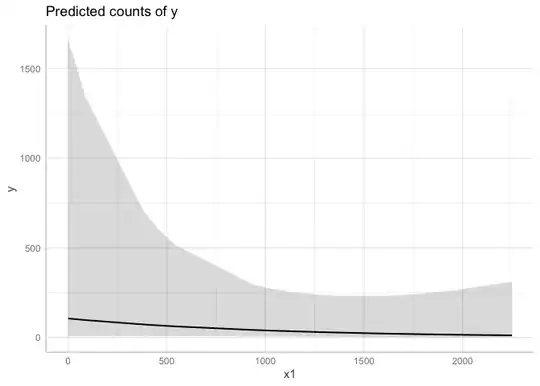
Is there any way to make a conditional plot using visreg fucntion when I have scaled my regressors? I want to plot my rescaled response along with 95%CIs, prediction curve, and partial residuals.
install.packages("lme4")
library("lme4")
y <- c(18, 0, 2, 0, 0, 0, 2, 0, 0, 1, 7, 0, 0, 0, 0, 0, 0, 0, 0)
x1 <- c(501, 1597, 1156, 1134, 1924, 507, 1022, 0, 92, 1729, 85, 963, 544, 1315, 2250, 1366, 458, 385, 930)
x2 <- c(0, 92, 959, 1146, 900, 0, 276, 210, 980, 8, 0, 473, 0, 255, 1194, 542, 983, 331, 923)
x3 <- c("site1", "site1", "site2","site2","site3","site3","site4","site4","site5","site5","site6","site7","site8","site9","site10","site11","site12","site13","site14")
offset_1 <- c(59, 34, 33, 35, 60, 58, 59, 33, 34, 61, 58, 58, 55, 26, 26, 18, 26, 26, 26)
data_1 <- data.frame(y,x1,x2,offset_1)
m1 <- glmer.nb(y ~ -1 + scale(x1) + scale(x2) + + (1|x3) + offset(log(offset_1)), data=data_1)
summary(m1)
visreg(m1, "x1", scale="response", cond=list(offset_1=1), partial=TRUE,
rug=2,line=list(lwd=0.5, col="black"), points=list(cex=1.4, lwd=0.1, col="black", pch=21))