Hi everyone I have this data:
structure(list(Site = c("aNon burn", "aNon burn", "aNon burn",
"Long rotation", "Long rotation", "Long rotation"), Depth = c("0-20cm",
"20-40cm", "40-60cm", "0-20cm", "20-40cm", "40-60cm"), bacteria = c(10.03,
8.46, 8.48, 9.34, 8.43, 8.45), bac_se = c(0.06, 0.06, 0.09, 0.08,
0.1, 0.05), fungi = c(8.18, 6.31, 6.45, 7.65, 5.88, 5.86), fun_se = c(0.21,
0.19, 0.17, 0.16, 0.17, 0.16)), row.names = c(NA, 6L), class = "data.frame")
and I would like to do a barplot with a single Y axis
I tried this code
ggplot(qpcrdata2, aes(x=Depth, y=bacteria, fill=Site)) + geom_bar(stat="identity", position=position_dodge()) +geom_errorbar(aes(ymin=bacteria-bac_se, ymax=bacteria+bac_se), width=.2,position=position_dodge(.9))
Gives me this figure
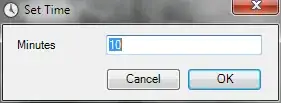
but I would also like to add Fungi on the same figure.
Can anyone help with this?
Best wishes