You can use package 'ggpmisc'. The functions in 'ggpubr' were based some years' back on those then existing in 'ggpmisc'. Since then many bugs were fixed and features added in 'ggpmisc' and the packages diverged. As the author of 'ggpmisc' I prefer it...
library(ggplot2)
library(ggpmisc)
#> Loading required package: ggpp
#>
#> Attaching package: 'ggpp'
#> The following object is masked from 'package:ggplot2':
#>
#> annotate
#> Registered S3 method overwritten by 'ggpmisc':
#> method from
#> as.character.polynomial polynom
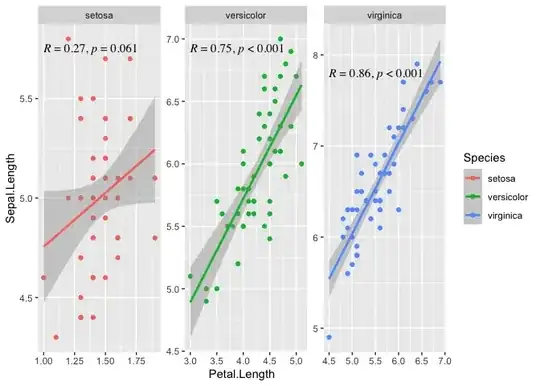
ggplot(data = iris,
aes(x = Petal.Length, y = Sepal.Length, color = Species))+
geom_point()+
geom_smooth(method = 'lm')+
facet_wrap('Species', scales = 'free') +
stat_correlation(use_label(c("R", "p")),
colour = "black",
p.digits = 3,
size = 3.5) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.12)))
#> `geom_smooth()` using formula = 'y ~ x'
Created on 2023-08-17 with reprex v2.0.2
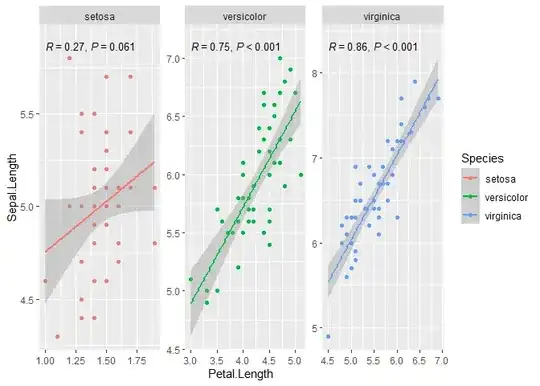
When there is a grouping, assuming that there is a single panel, we need to displace the labels for each group vertically so that they do not overlap. In stat_correlation() vstep controls how big this displacement is, so setting it to zero solves this problem, if we want to keep the colour mapping for the labels (e.g., replace colour = "black" with vstep = 0). I expanded the y scale so that the labels do not overlap with the observations. I forgot to add family = 'serif' and I put a slightly smaller size. Of course, this can changed to match your example.
stat_correlation() can do a few more tricks, such computing the confidence interval for R, rho or tau, or use r instead of R, and p instead of P if preferred. There are examples in the documentation of 'ggpmisc' and at https://www.r4photobiology.info/galleries/plot-equations.html#linear-correlation