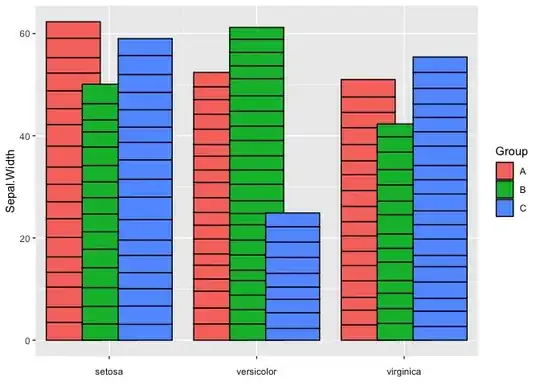
As already suggested by @r2evans in his comment one option to fake dodging and stacking in the same chart would be to use the "facets that do not look like facets" trick, which means to facet by Species and to map Group on x. Additionally, to achieve the overlap of the stacked bars for the three groups one could set the width of the bars to a value > 1, which also requires to adjust the limits (which I do via coord_cartesian) and to arrange your data by Group. Finally, we have to do some adjustments via theme() to get rid of the facet look.
set.seed(101)
x <- cbind(iris,
Group = sample(LETTERS[1:3], size = nrow(iris), replace = TRUE)
)
library(ggplot2)
library(dplyr, warn = FALSE)
x |>
dplyr::arrange(Species, Group) |>
ggplot() +
geom_col(
aes(x = Group, y = Sepal.Width, fill = Group),
col = 1, width = 1.5
) +
scale_x_discrete(breaks = "B") +
facet_wrap(~Species, scales = "free_x", strip.position = "bottom") +
coord_cartesian(xlim = c(.55, 3.45)) +
theme(
axis.text.x = element_blank(),
strip.background.x = element_blank(),
panel.spacing.x = unit(0, "pt"),
strip.placement = "outside"
) +
labs(x = NULL)
#> Warning: `position_stack()` requires non-overlapping x intervals
#> `position_stack()` requires non-overlapping x intervals
#> `position_stack()` requires non-overlapping x intervals